Visualization for Machine Learning
Spring 2024
Agenda
- Clustering
- Dimensionality Reduction
Clustering
Etienne Bernard: “… the goal of clustering is to separate a set of examples into groups called clusters”
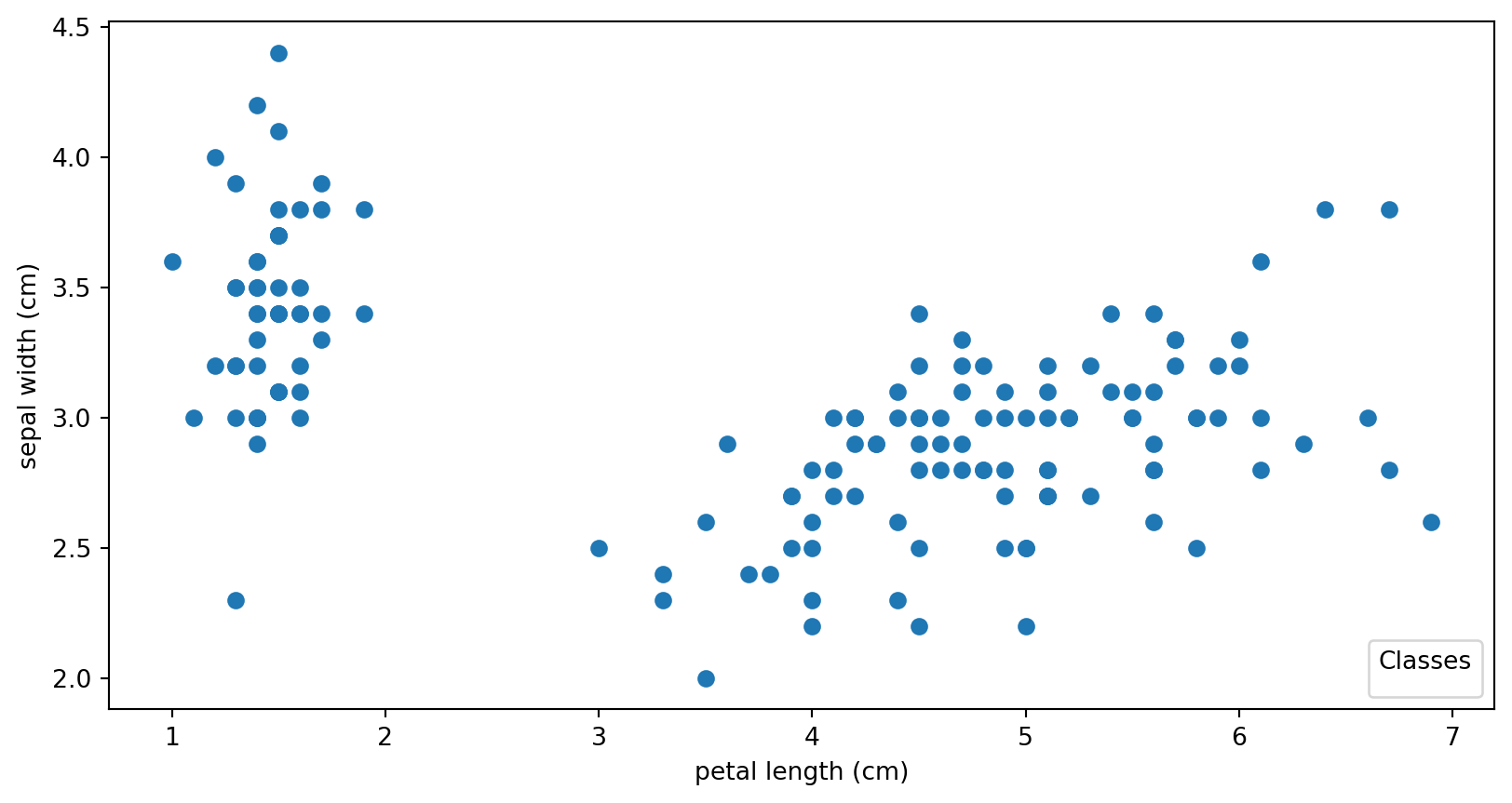
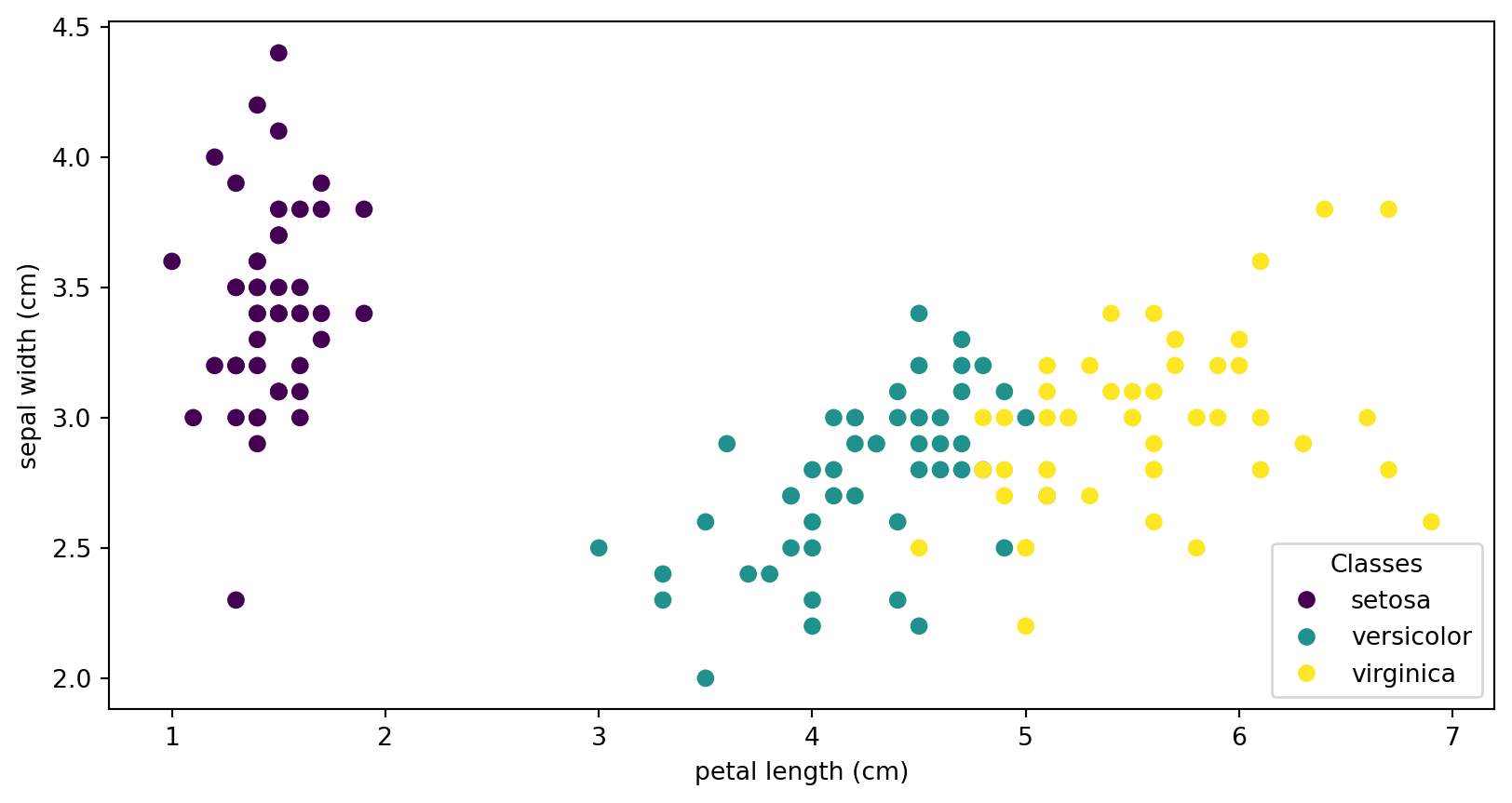
IRIS
# Code source: Gaël Varoquaux
# Modified for documentation by Jaques Grobler
# License: BSD 3 clause
#
import matplotlib.pyplot as plt
from sklearn import datasets
iris = datasets.load_iris()
_, ax = plt.subplots()
scatter = ax.scatter(iris.data[:, 2], iris.data[:, 1])
ax.set(xlabel=iris.feature_names[2], ylabel=iris.feature_names[1])
_ = ax.legend(
scatter.legend_elements()[0], iris.target_names, loc="lower right", title="Classes"
)IRIS
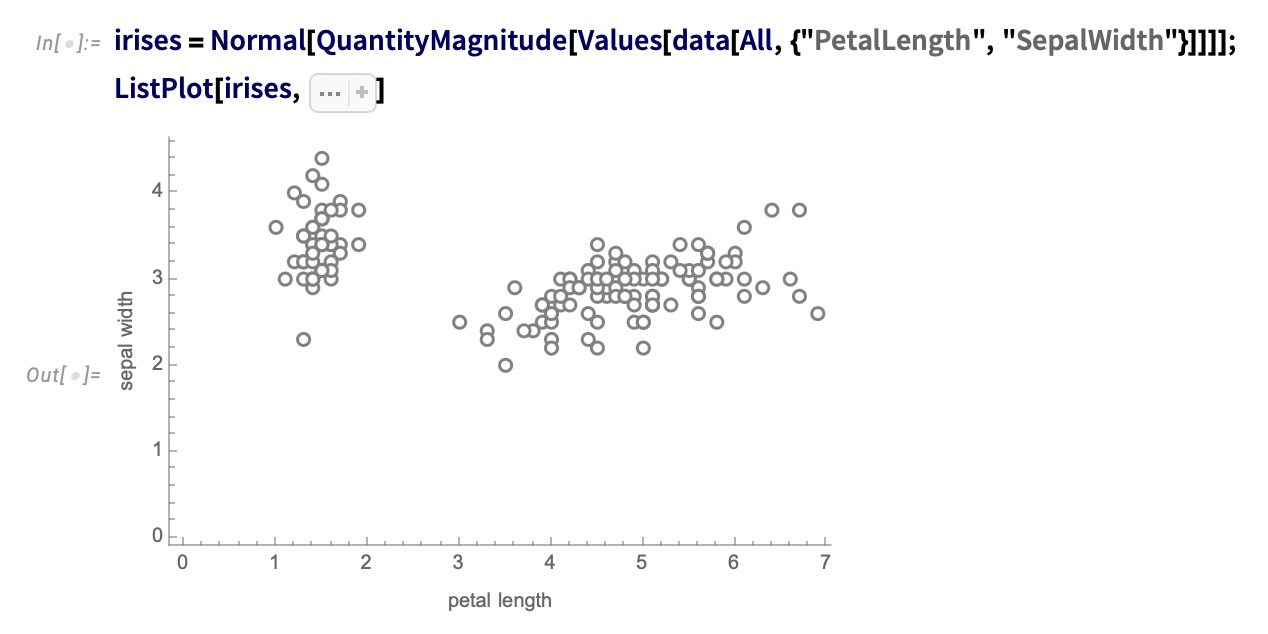
IRIS – another look (Bernard)
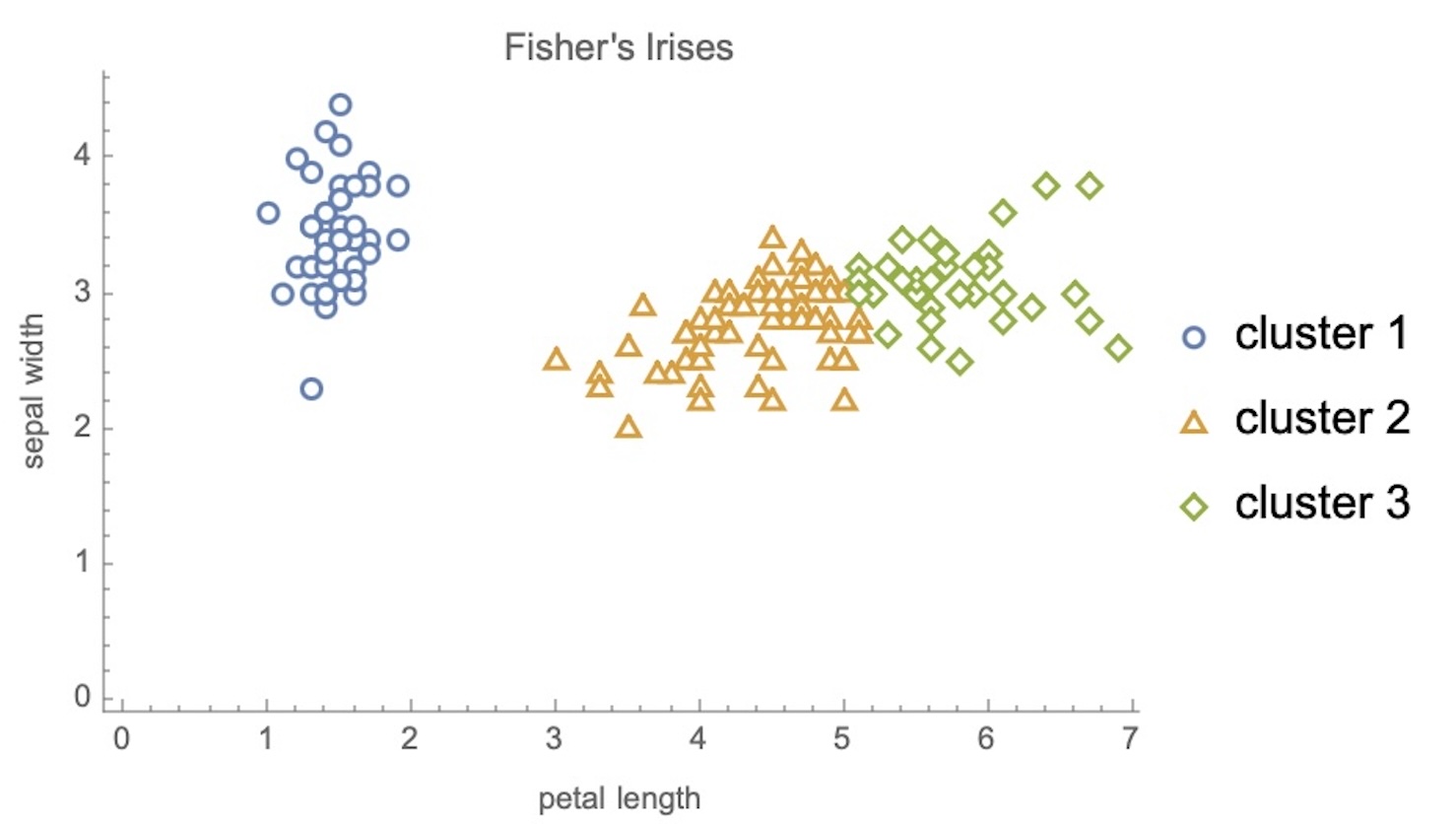
IRIS – clustering
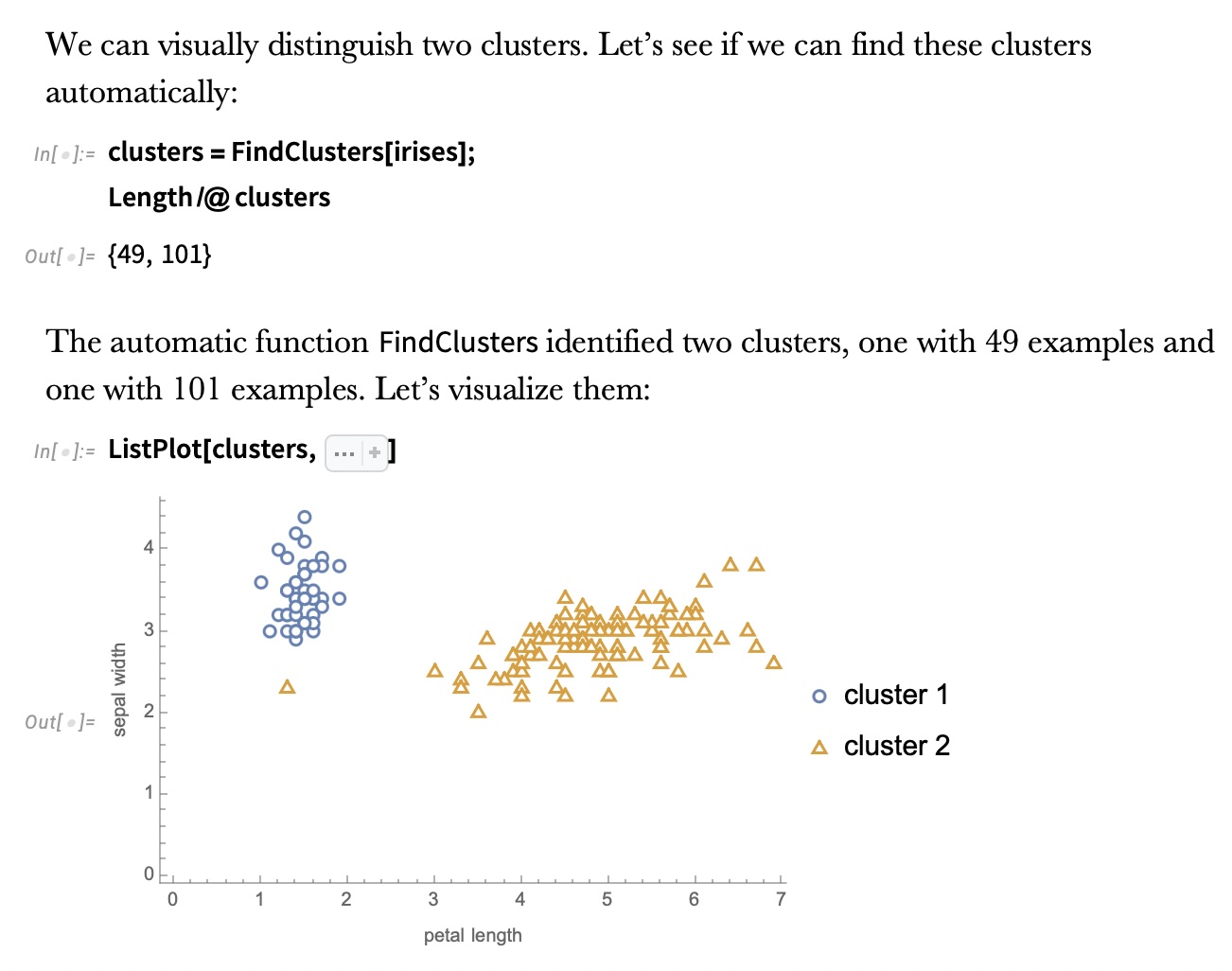
IRIS – k-means
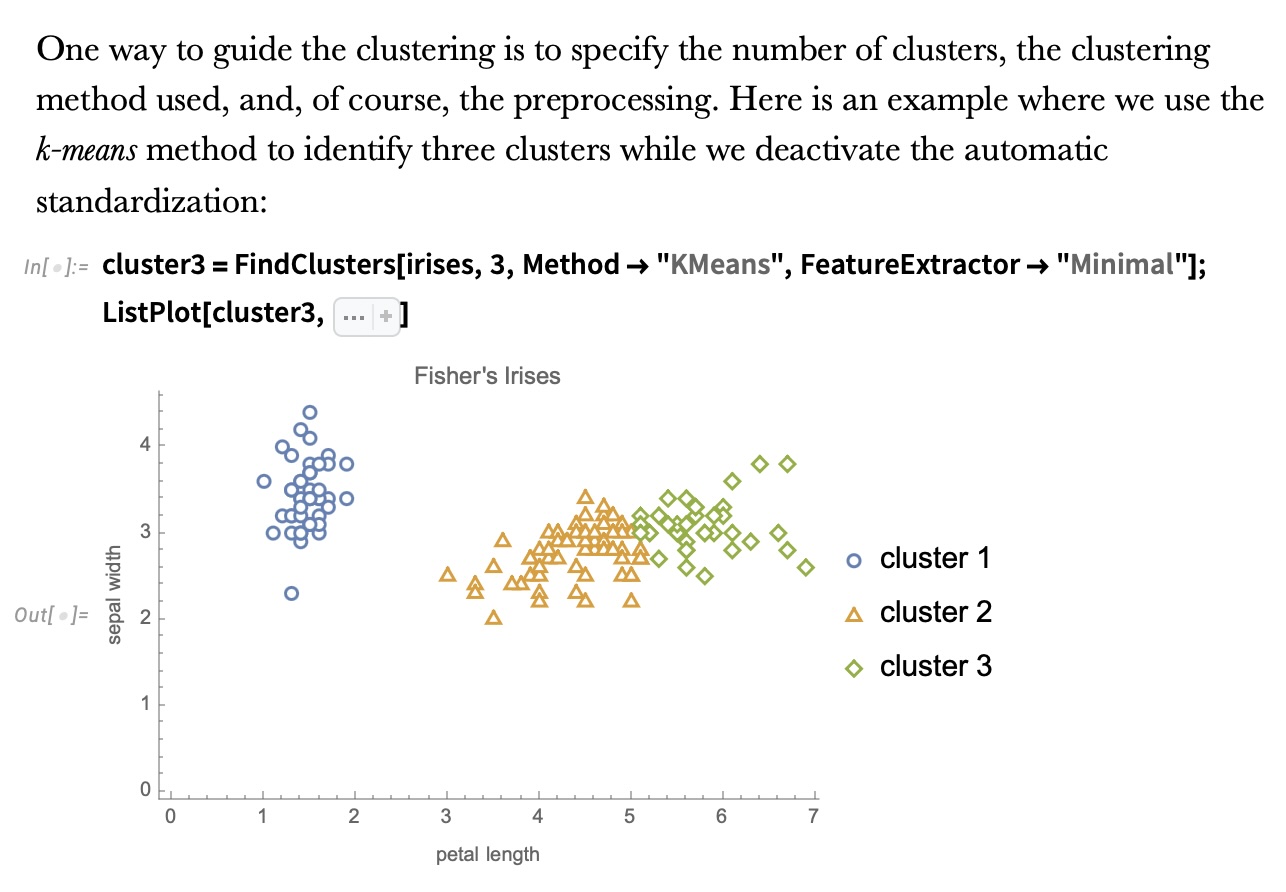
Wolfram Mathematica FindClusters
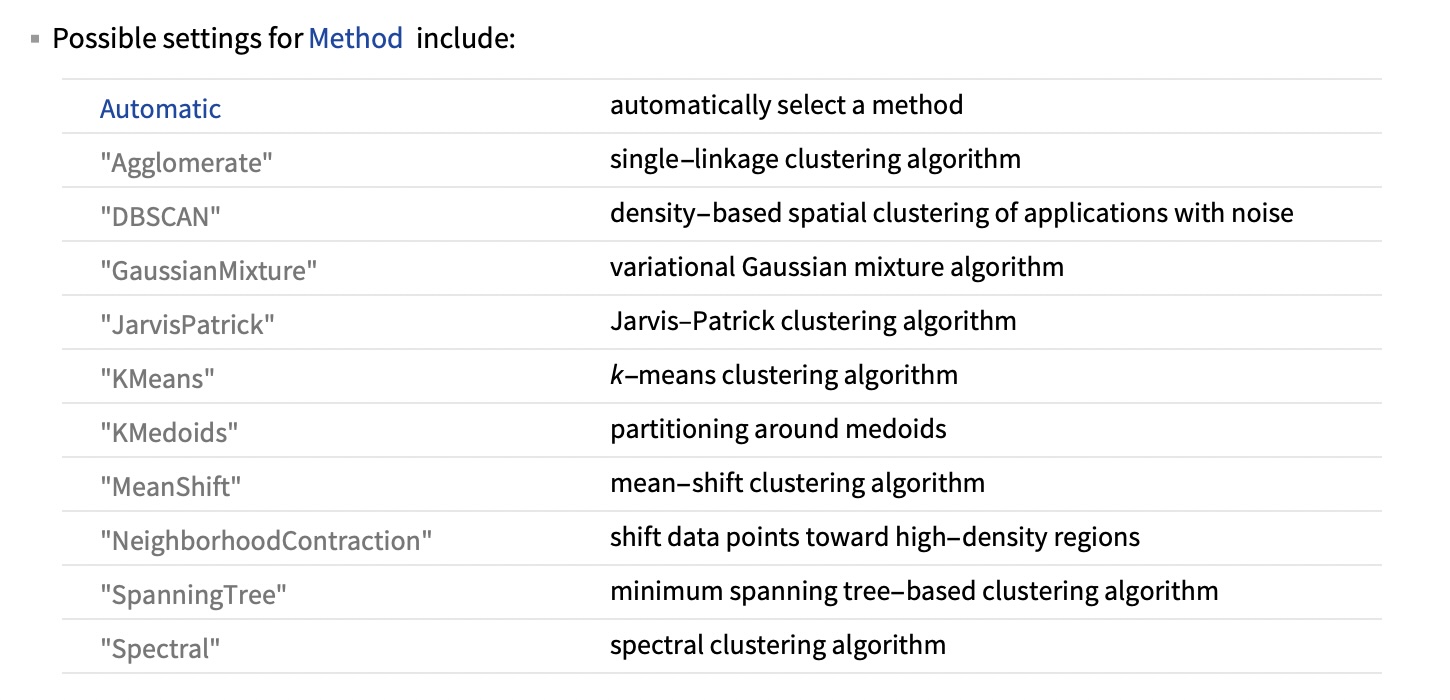
Wolfram Mathematica FindClusters
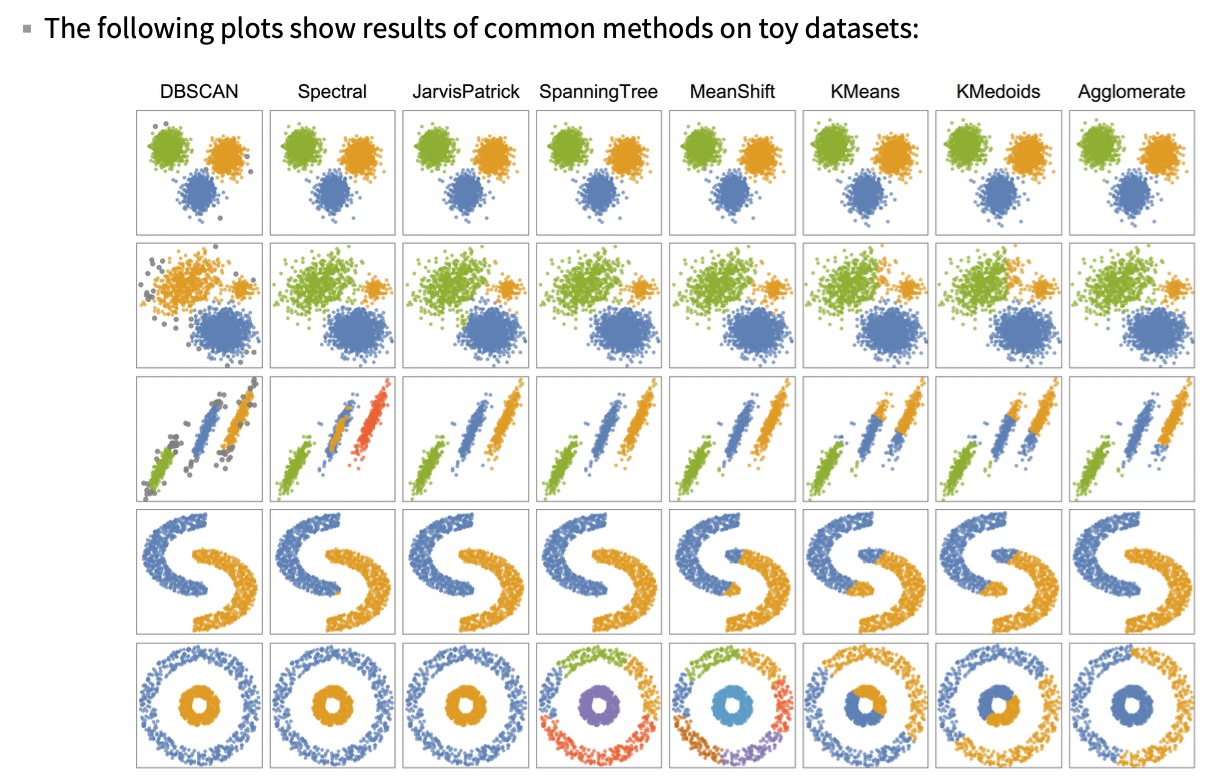
IRIS - classes
Recommended reading
Required https://www.wolfram.com/language/introduction-machine-learning/clustering/ link
https://en.wikipedia.org/wiki/Cluster_analysis
https://en.wikipedia.org/wiki/K-means_clustering
https://en.wikipedia.org/wiki/DBSCAN
Dimensionality Reduction
Input data may have thousands or millions of dimensions!
Dimensionality Reduction represents data with fewer dimensions
- easier learning – fewer parameters
- visualization – show high-dimensional data in 2D or 3D
- discover “intrinsic dimensionality” of the data
Dimensionality Reduction (Yi Zhang)
- Assumption: data lies on a lower dimensional space
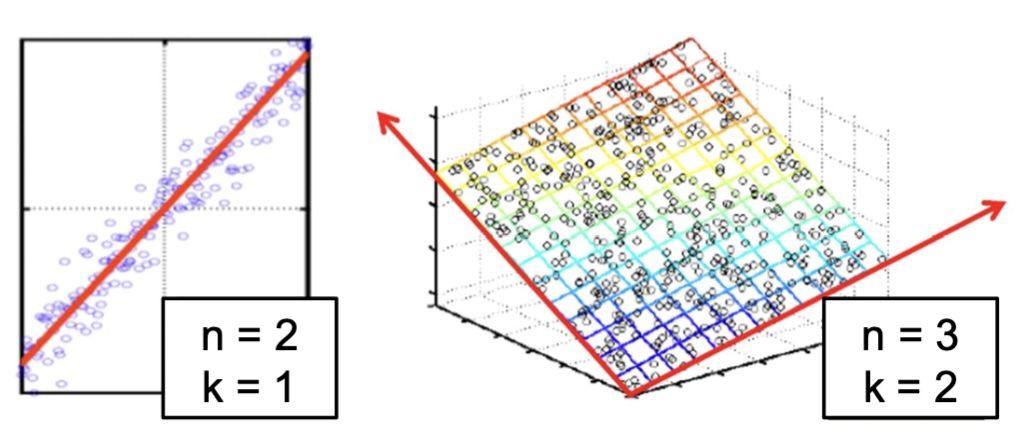
Dimensionality Reduction (Bishop)
- Supposed a dataset of “3s” perturbed in various ways
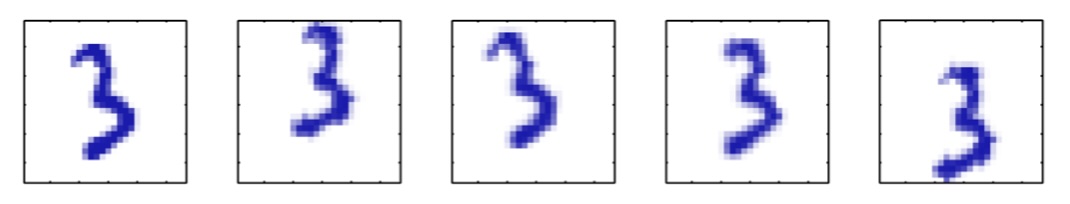
What operations did we perform? What’s the intrinsic dimensionality?
Here the underlying manifold is non-linear
Digits
Digits - 0
array([[ 0., 0., 5., 13., 9., 1., 0., 0.],
[ 0., 0., 13., 15., 10., 15., 5., 0.],
[ 0., 3., 15., 2., 0., 11., 8., 0.],
[ 0., 4., 12., 0., 0., 8., 8., 0.],
[ 0., 5., 8., 0., 0., 9., 8., 0.],
[ 0., 4., 11., 0., 1., 12., 7., 0.],
[ 0., 2., 14., 5., 10., 12., 0., 0.],
[ 0., 0., 6., 13., 10., 0., 0., 0.]])Digits - 1
array([[ 0., 0., 0., 12., 13., 5., 0., 0.],
[ 0., 0., 0., 11., 16., 9., 0., 0.],
[ 0., 0., 3., 15., 16., 6., 0., 0.],
[ 0., 7., 15., 16., 16., 2., 0., 0.],
[ 0., 0., 1., 16., 16., 3., 0., 0.],
[ 0., 0., 1., 16., 16., 6., 0., 0.],
[ 0., 0., 1., 16., 16., 6., 0., 0.],
[ 0., 0., 0., 11., 16., 10., 0., 0.]])Digits
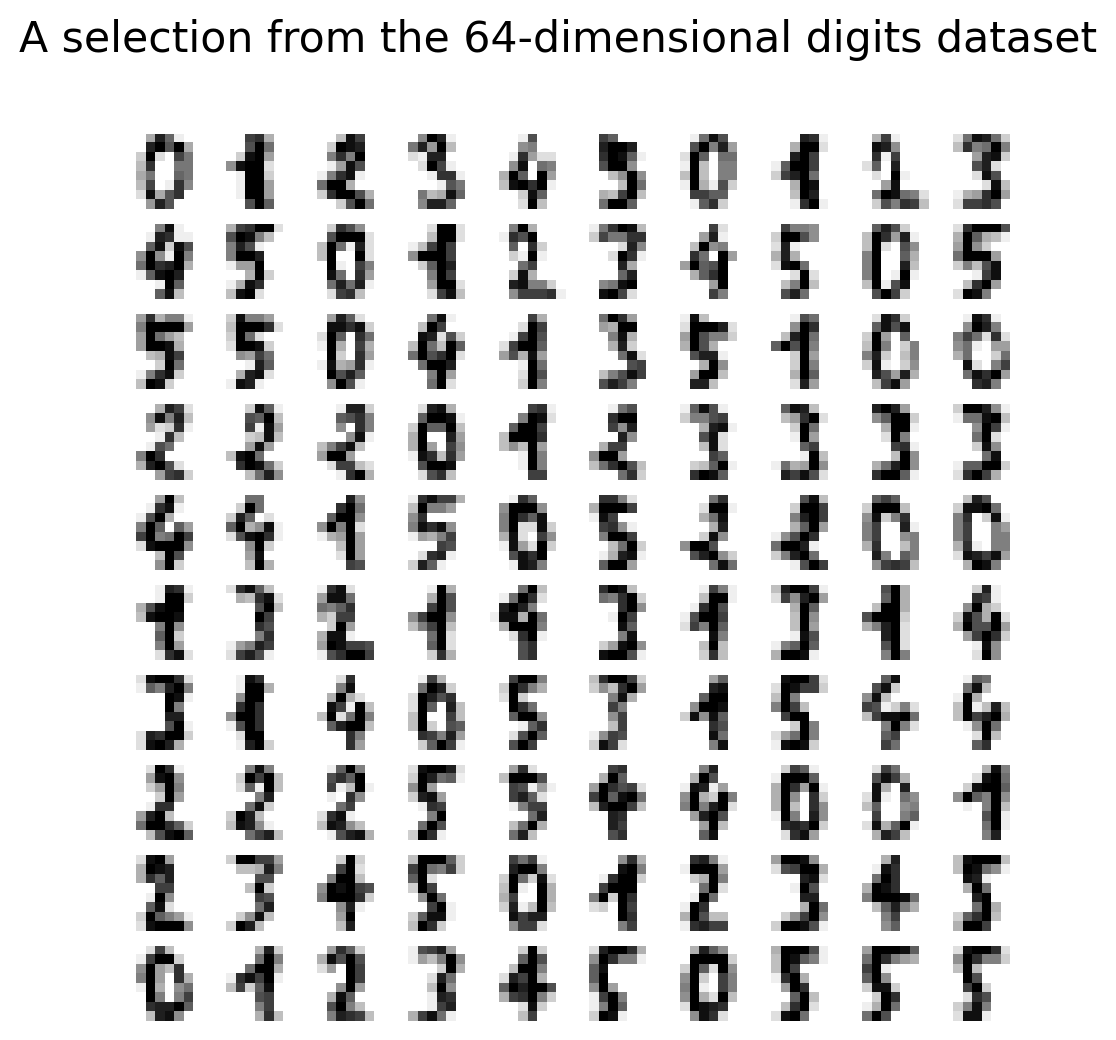
Digits
Computing Random projection embedding...
Computing Truncated SVD embedding...
Computing Linear Discriminant Analysis embedding...
Computing Isomap embedding...
Computing Standard LLE embedding...
Computing Modified LLE embedding...
Computing Hessian LLE embedding...
Computing LTSA LLE embedding...
Computing MDS embedding...
Computing Random Trees embedding...
Computing Spectral embedding...
Computing t-SNE embedding...
Computing NCA embedding...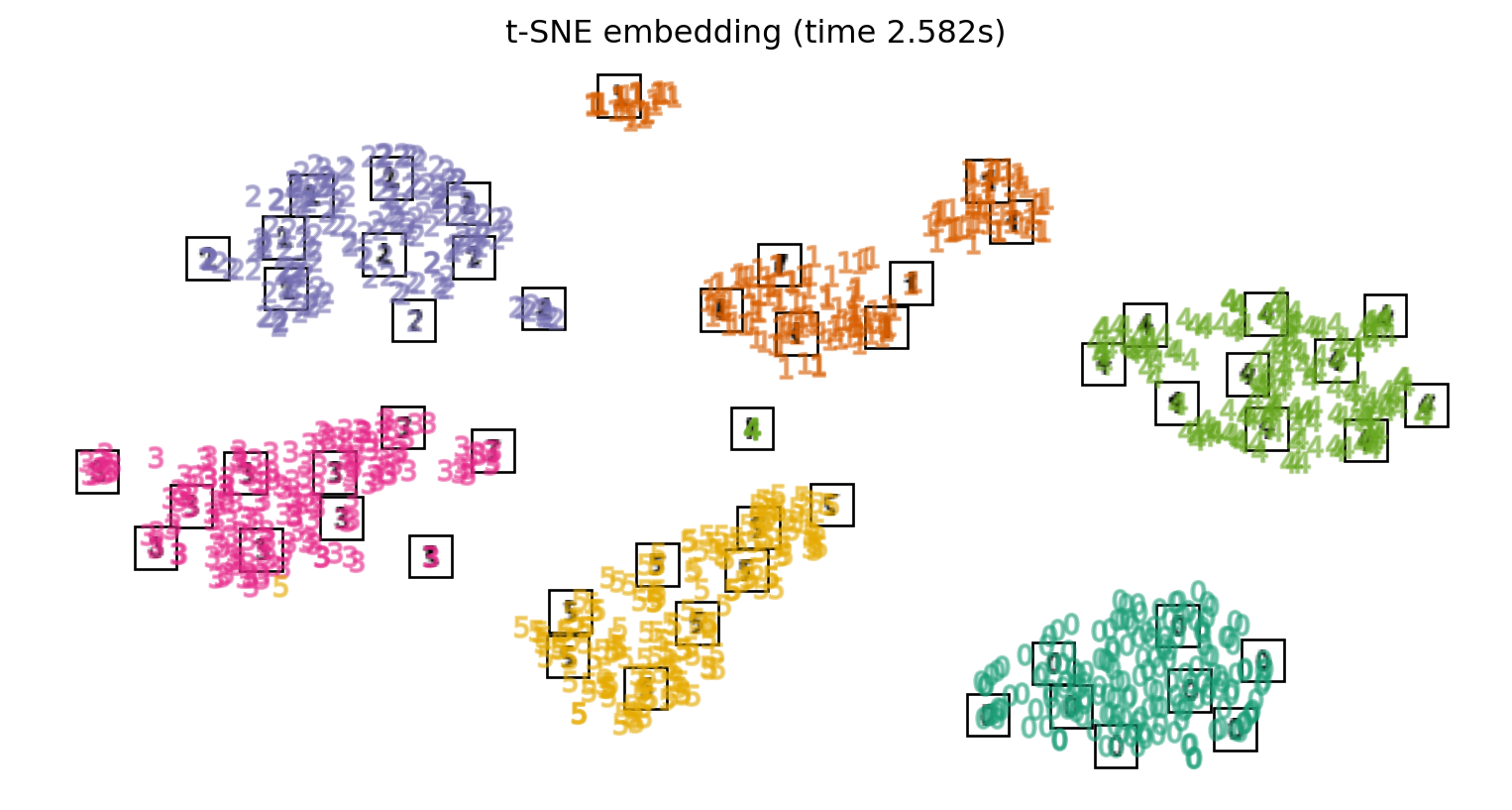
Principal Component Analysis (Luis Gustavo Nonato)
- PCA is directly related to the eigenvectors and eigenvalues of covariance matrices.
- Lets so make a quick review of eigenvectors, eigenvalues, and covariance matrices.
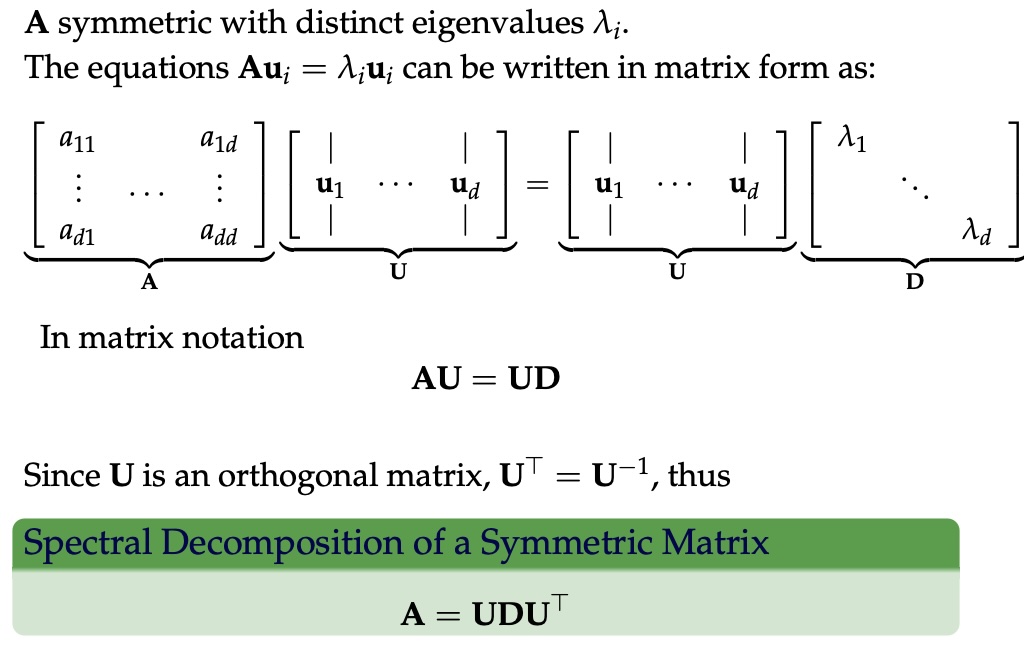
Eigenvectors and Eigenvalues
Given a \(d \times d\) matrix \(A\), a pair \((\lambda, u)\) that satisfies
\(A u = \lambda u\)
is called eigenvalue \(\lambda\) and corresponding eigenvector \(u\) of \(A\).
Symmetric Matrices
- \(\lambda \in \mathbb{R}\) and \(u \in \mathbb{R}^d\) (no complex numbers involved)
- The eigenvectors are orthogonal
Covariance Matrix

Covariance Matrix
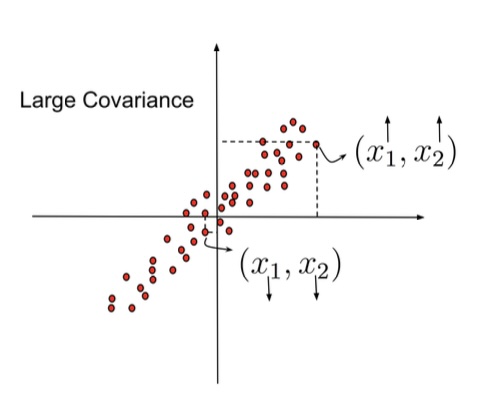
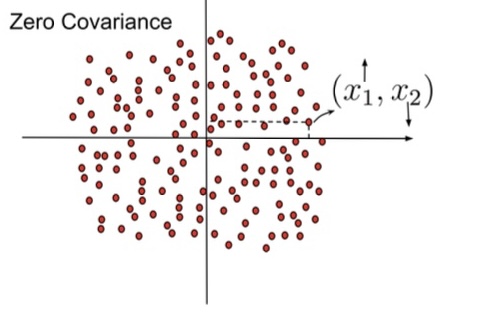
Covariance Matrix
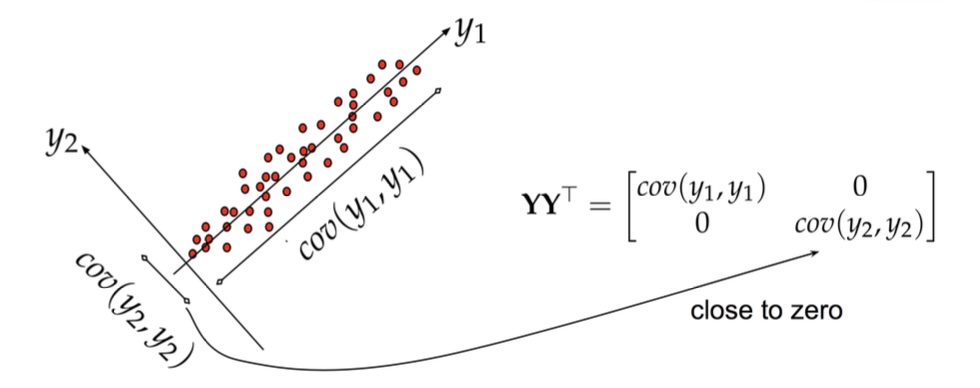
Principal Component Analysis: intuition
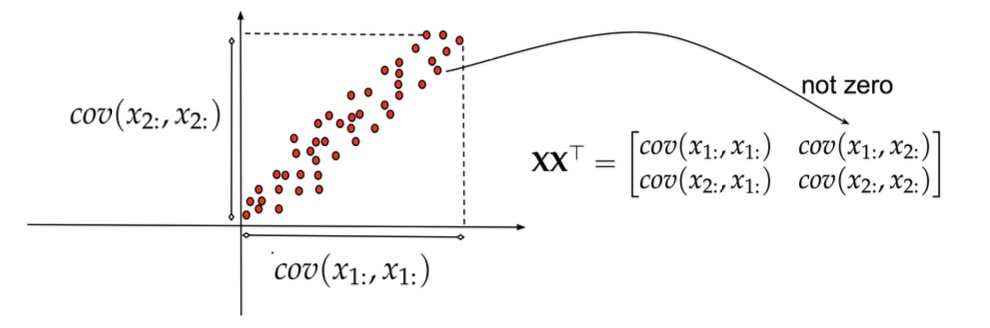
Principal Component Analysis: intuition
Principal Component Analysis

Principal Component Analysis
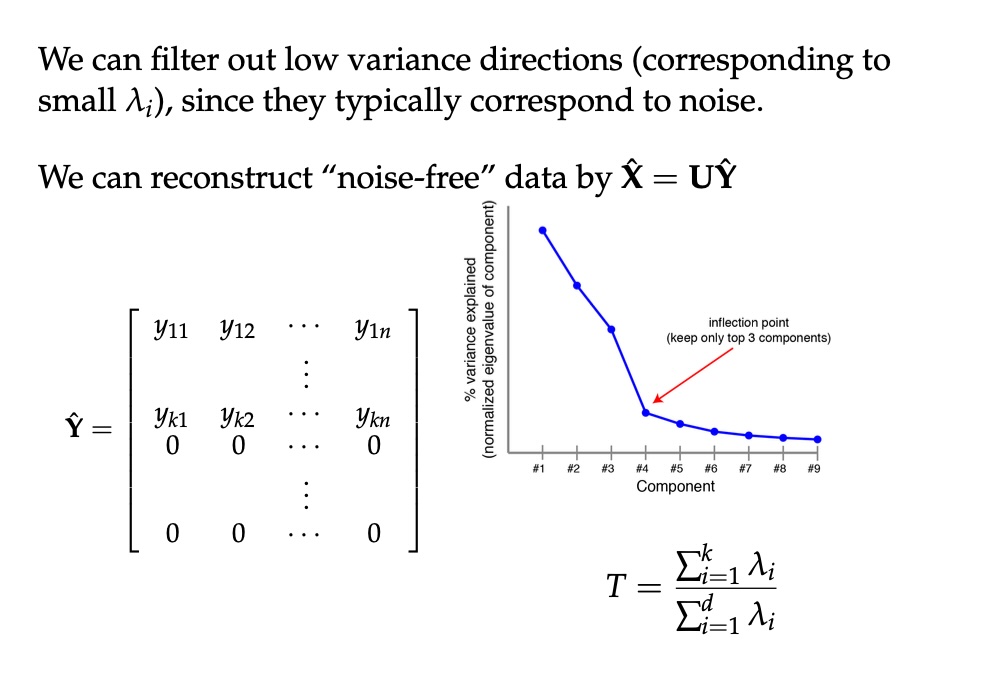
PCA of digits
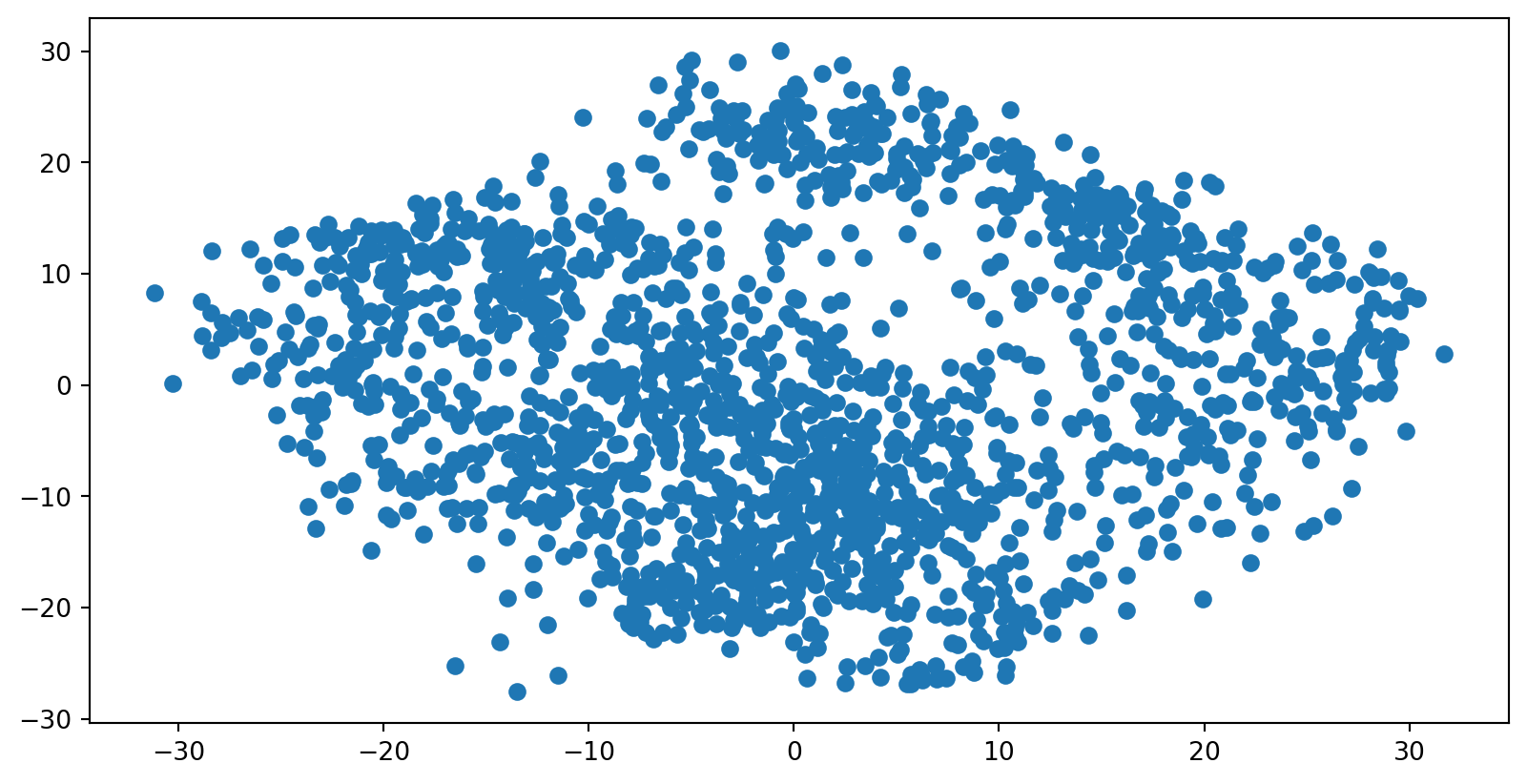
PCA of digits
Scaling Up
- Covariance matrix can be really big!
- \(\Sigma\) is \(n\) by \(n\)
- 10000 features are not uncommon
- computing eigenvectors is slow…
- Solution: Singular Value Decomposition (SVD)
- Finds the \(k\) largest eigenvectors
- Widely implemented robustly in major packages
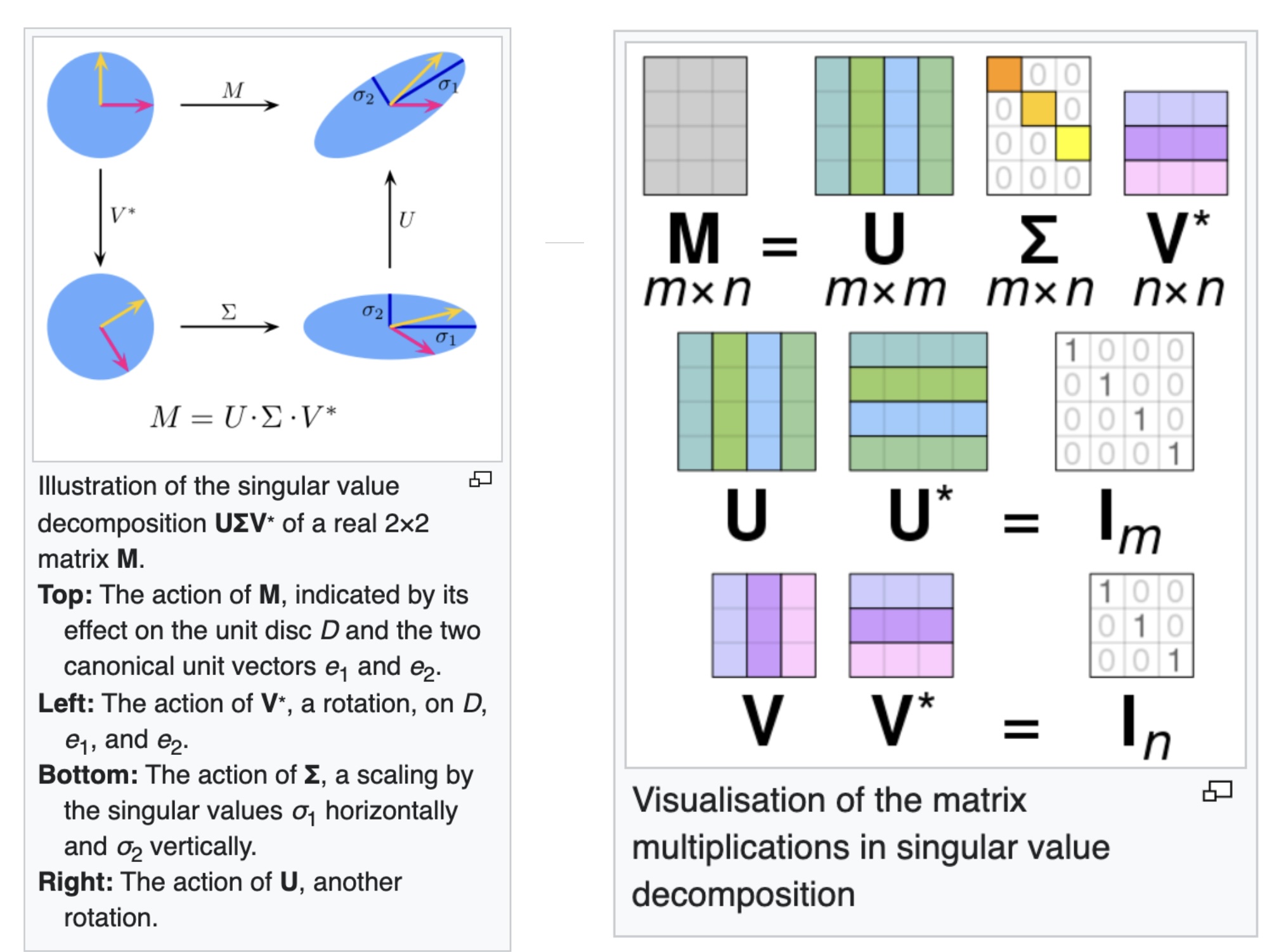
Singular Value Decomposition (SVD)
- https://en.wikipedia.org/wiki/Singular_value_decomposition
Dimensionality Reduction Techniques
- https://en.wikipedia.org/wiki/Dimensionality_reduction
- Principal component analysis (PCA)
- Non-negative matrix factorization (NMF)
- Linear discriminant analysis (LDA)
- t-SNE
- UMAP
- many others
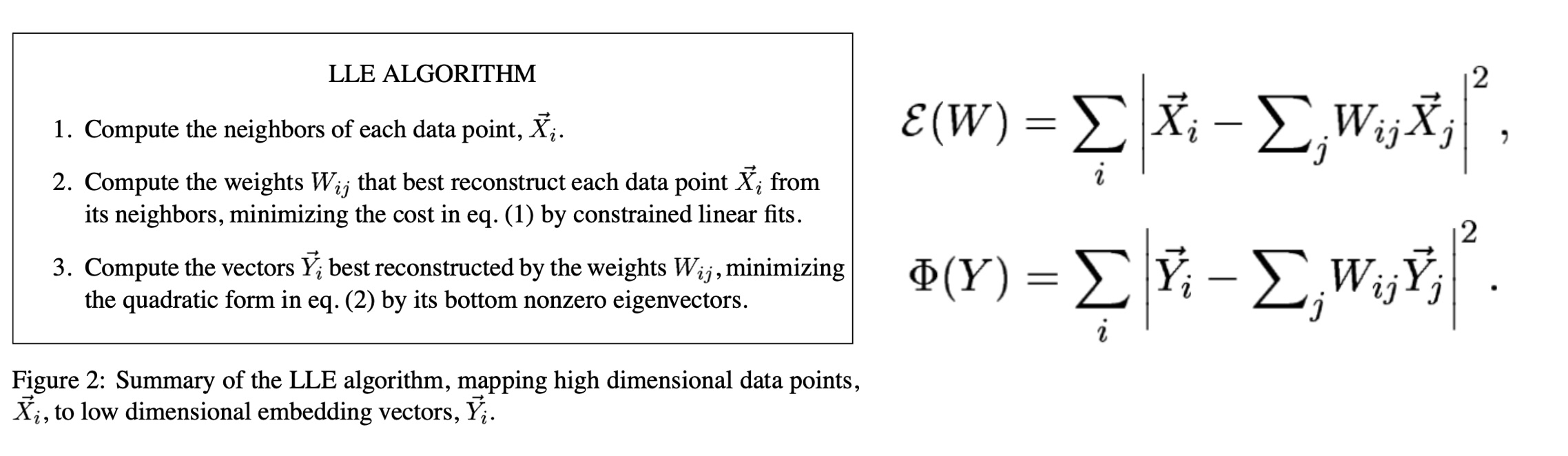
Local Linear Embedding
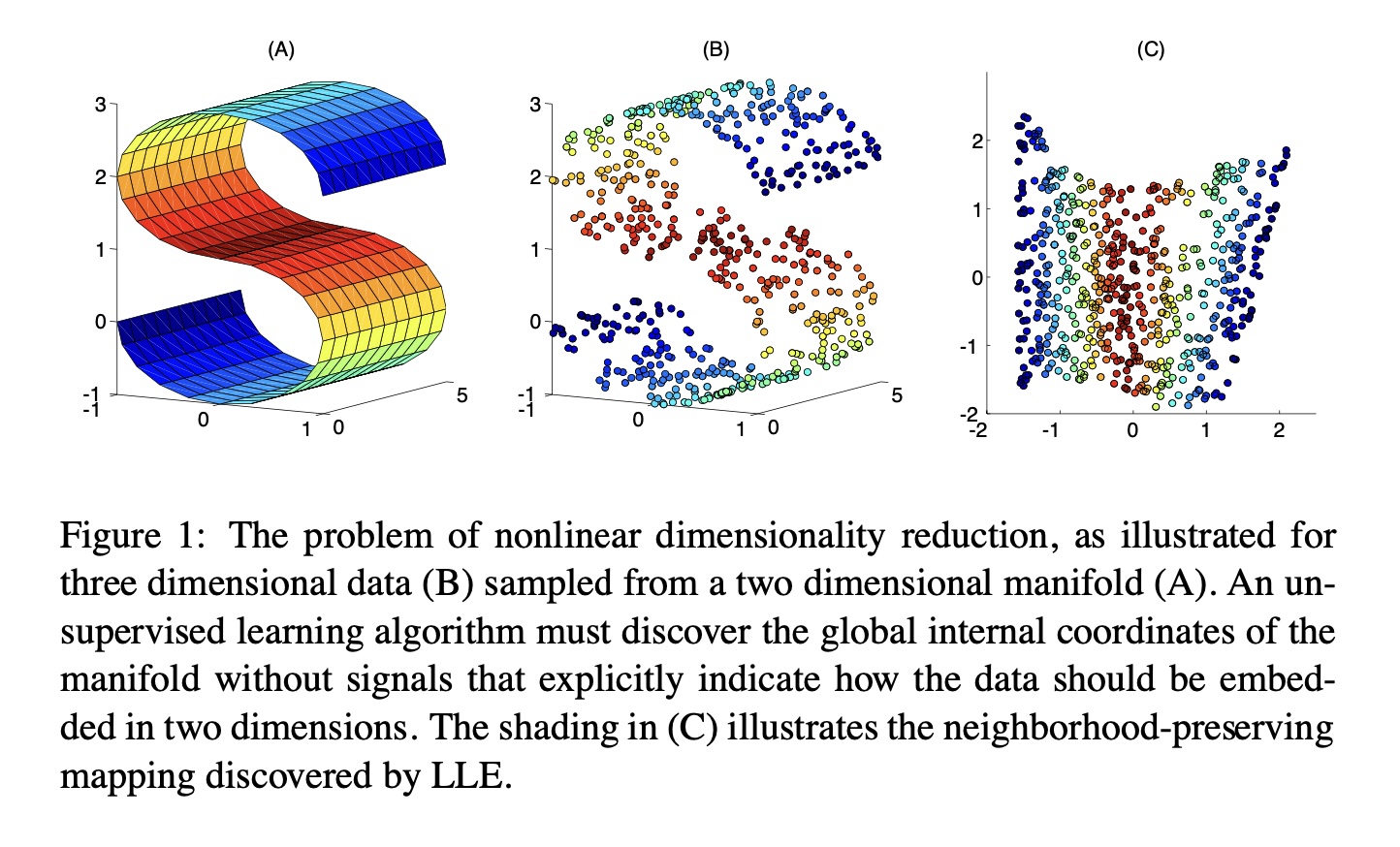
Preserving Local Manifold Neighborhoods
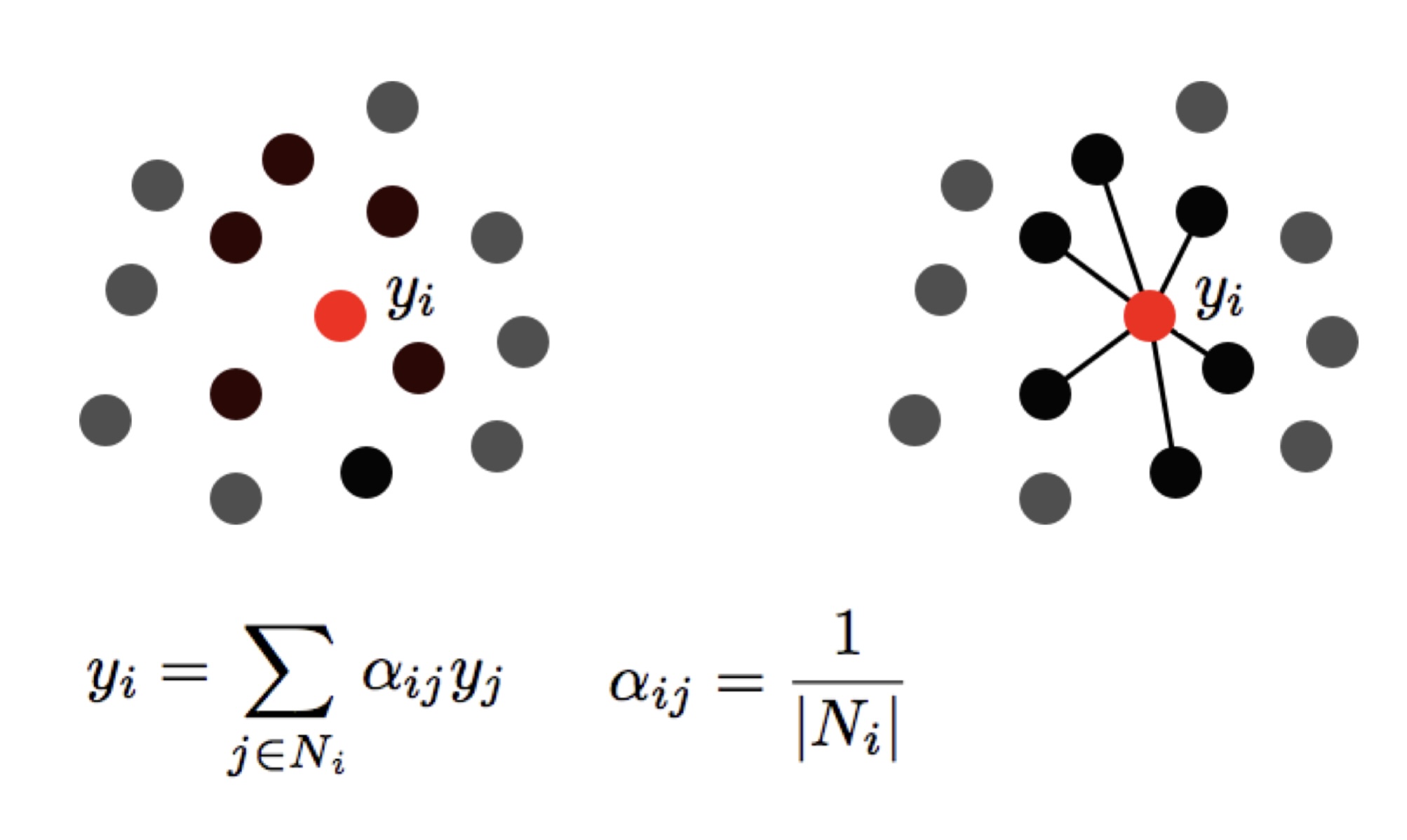
LLE
https://www.science.org/doi/10.1126/science.290.5500.2323
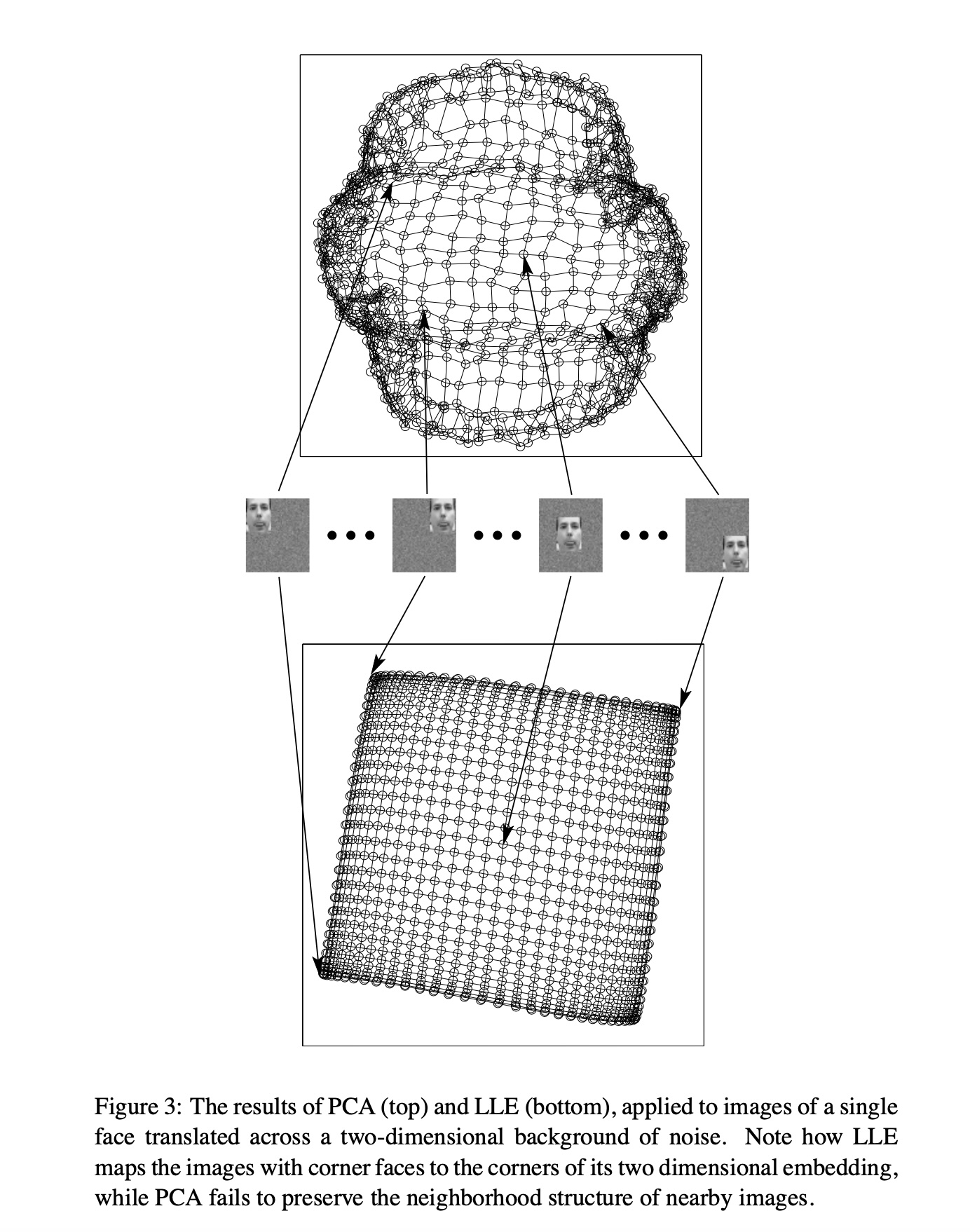
PCA vs LLE
Graph Layout using force based approach
SNE and t-SNE

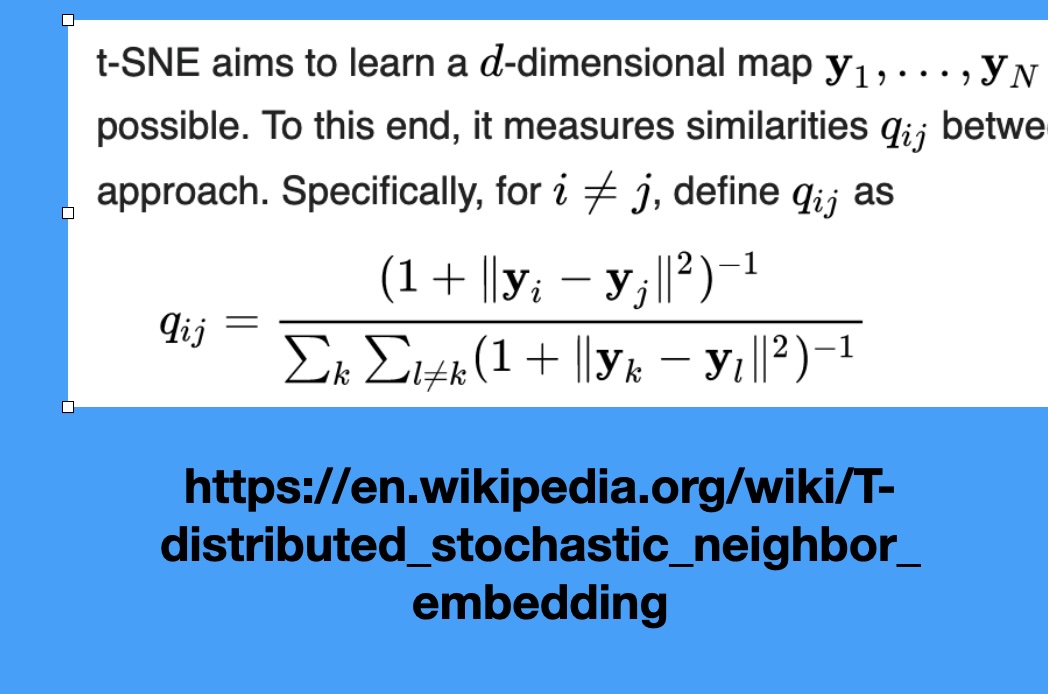
HERE is an excellent talk by t-SNE creator: video link
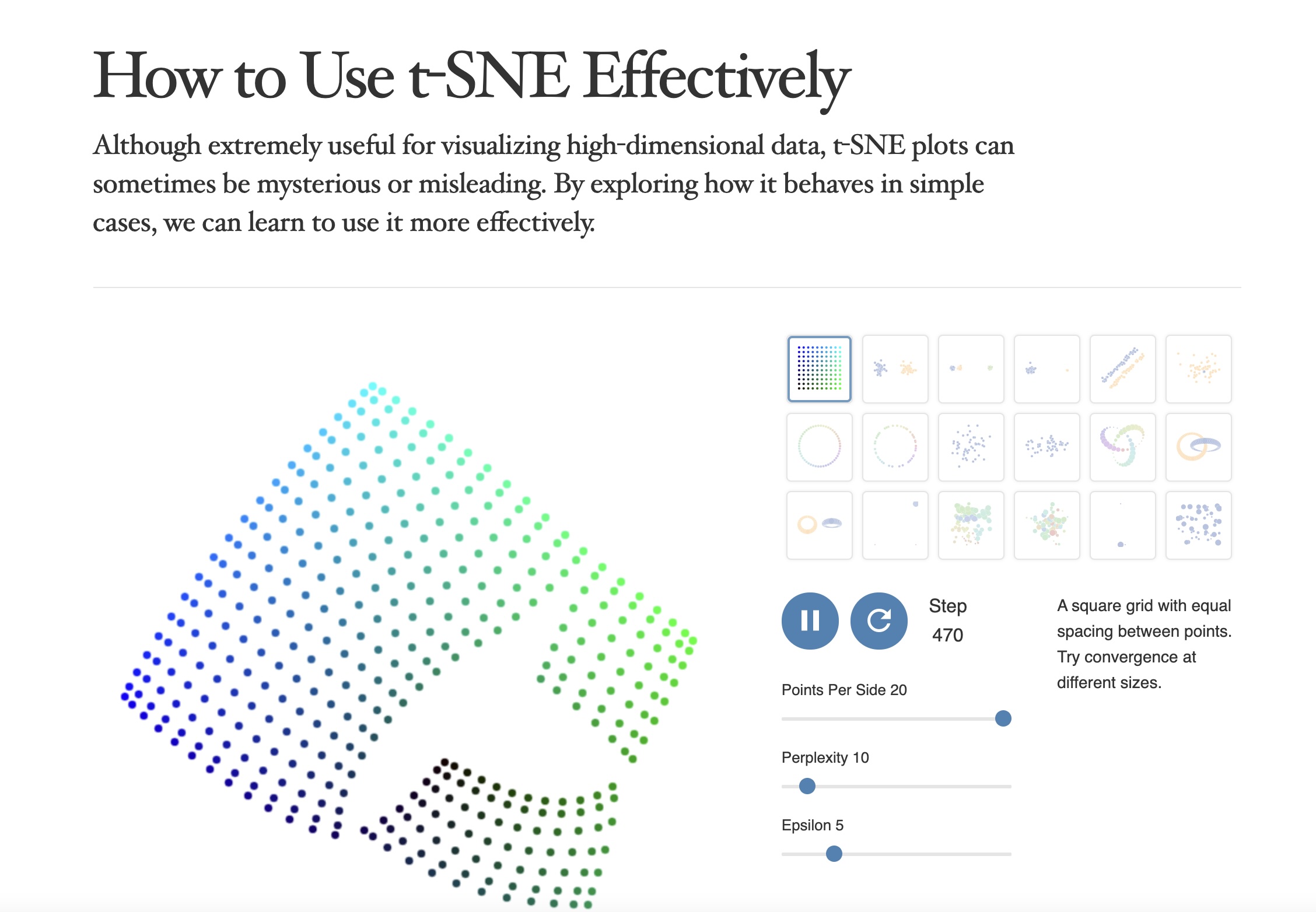
https://distill.pub/2016/misread-tsne/
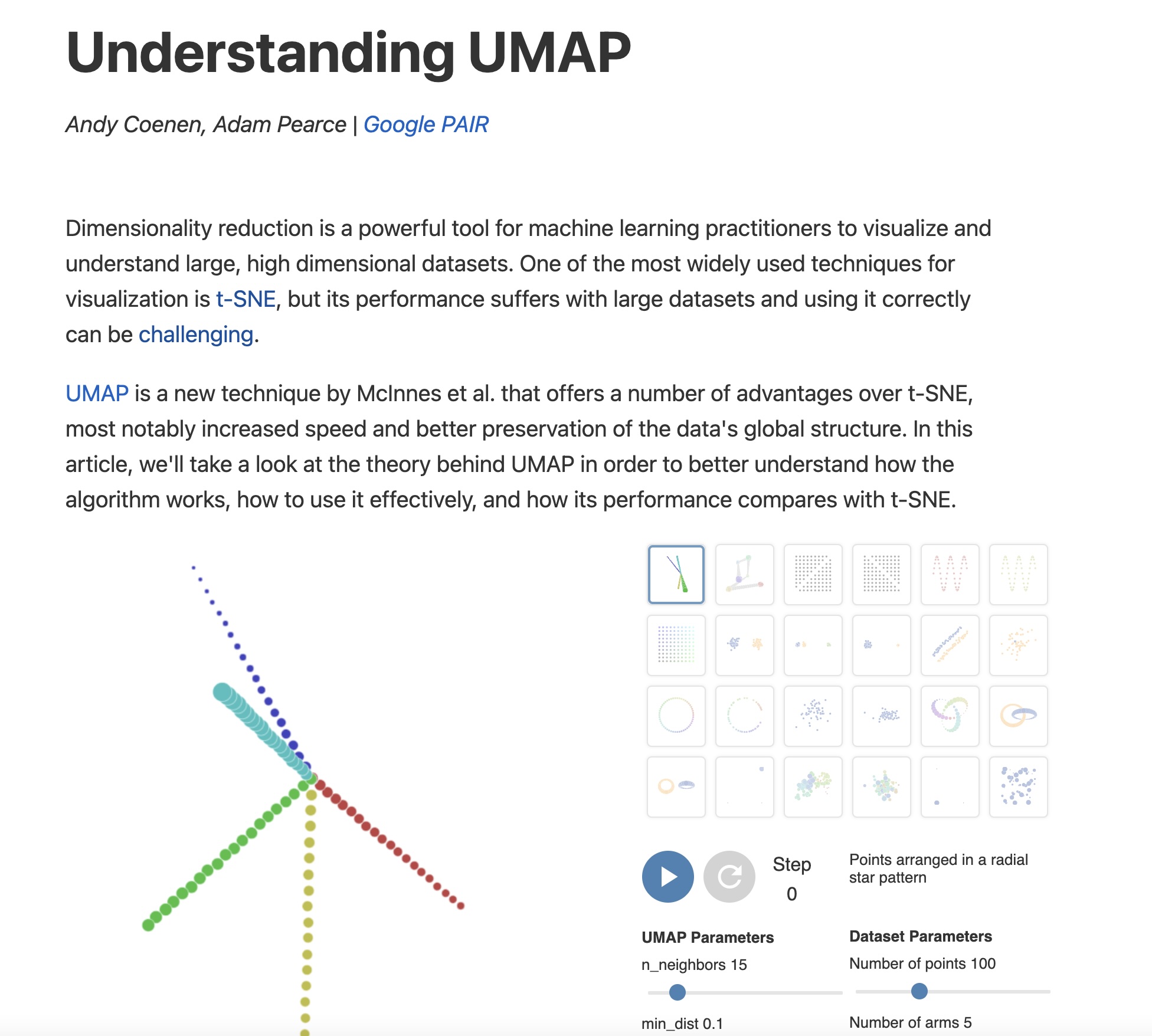
https://pair-code.github.io/understanding-umap/
What about user interaction?