Clustering and Dimensionality Reduction
CS-GY 6313 - Fall 2025
Claudio Silva
Agenda
- Clustering Visualization
- Introduction to clustering analysis
- K-means and visual patterns
- Comparing clustering methods
- Dimensionality Reduction for Visualization
- Why reduce dimensions?
- Linear methods: PCA
- Non-linear methods: t-SNE and UMAP
- Critical visualization principles
Clustering Visualization
What is Clustering?
“… the goal of clustering is to separate a set of examples into groups called clusters”
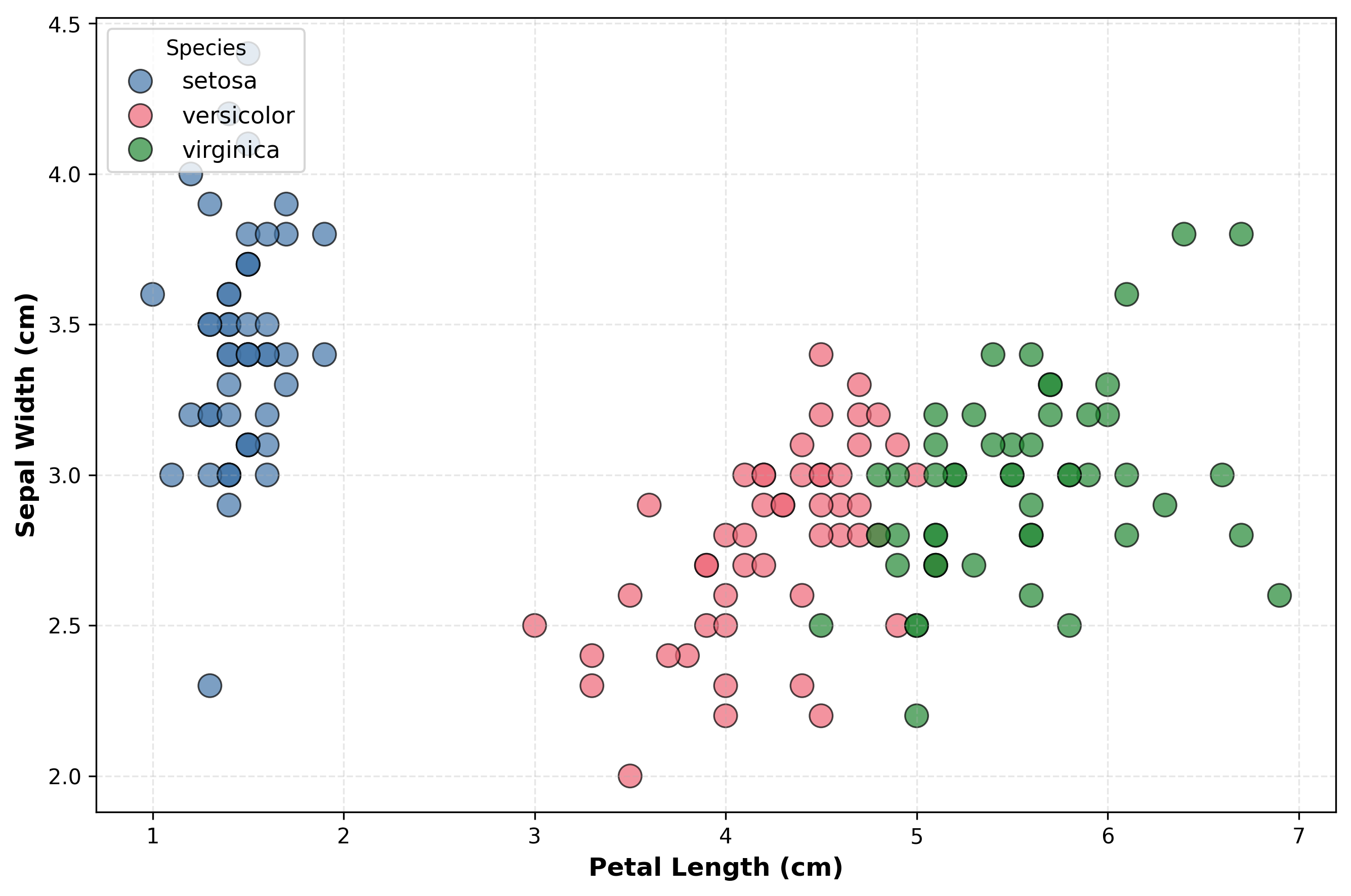
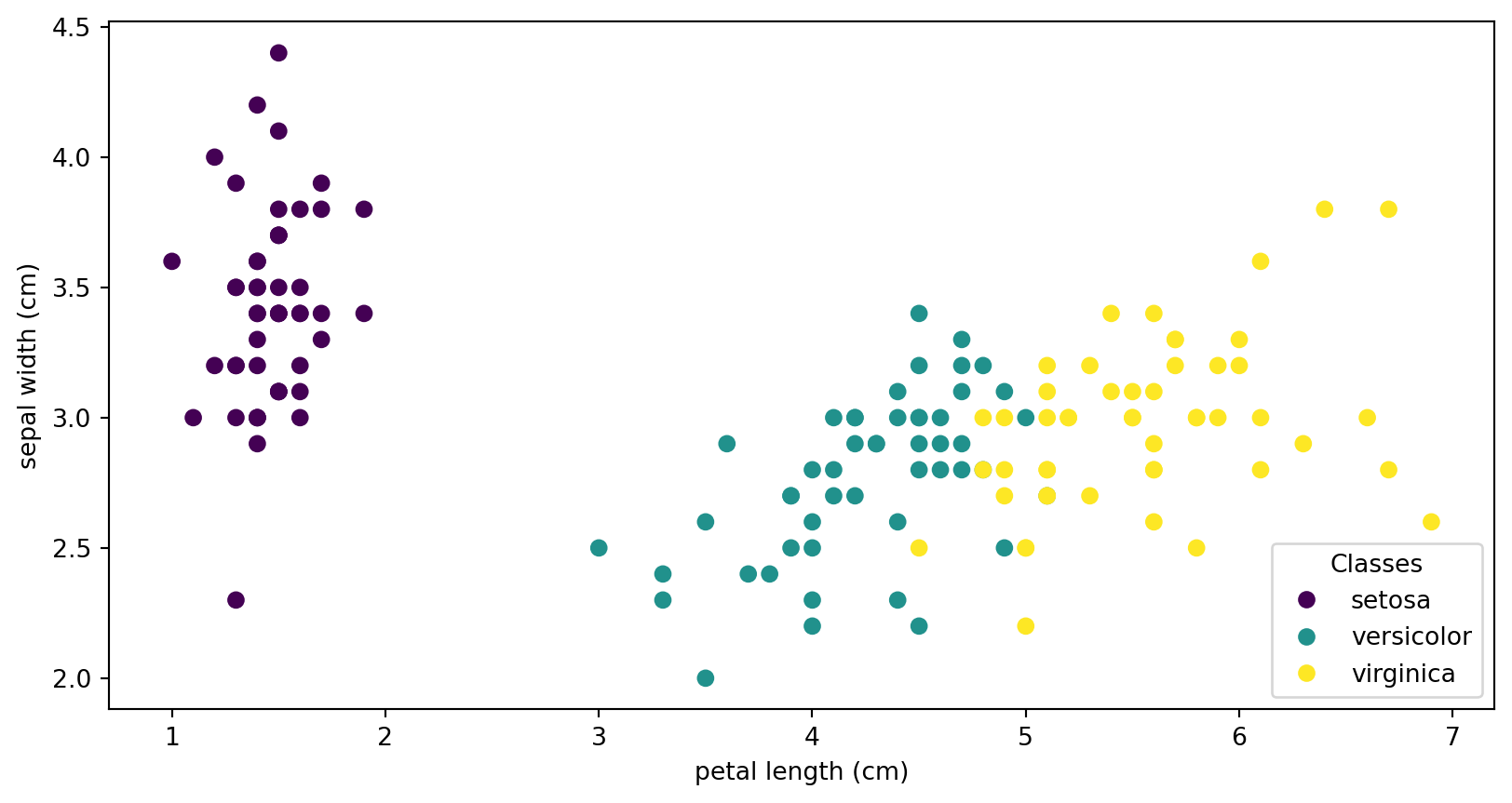
Visualizing the IRIS Dataset
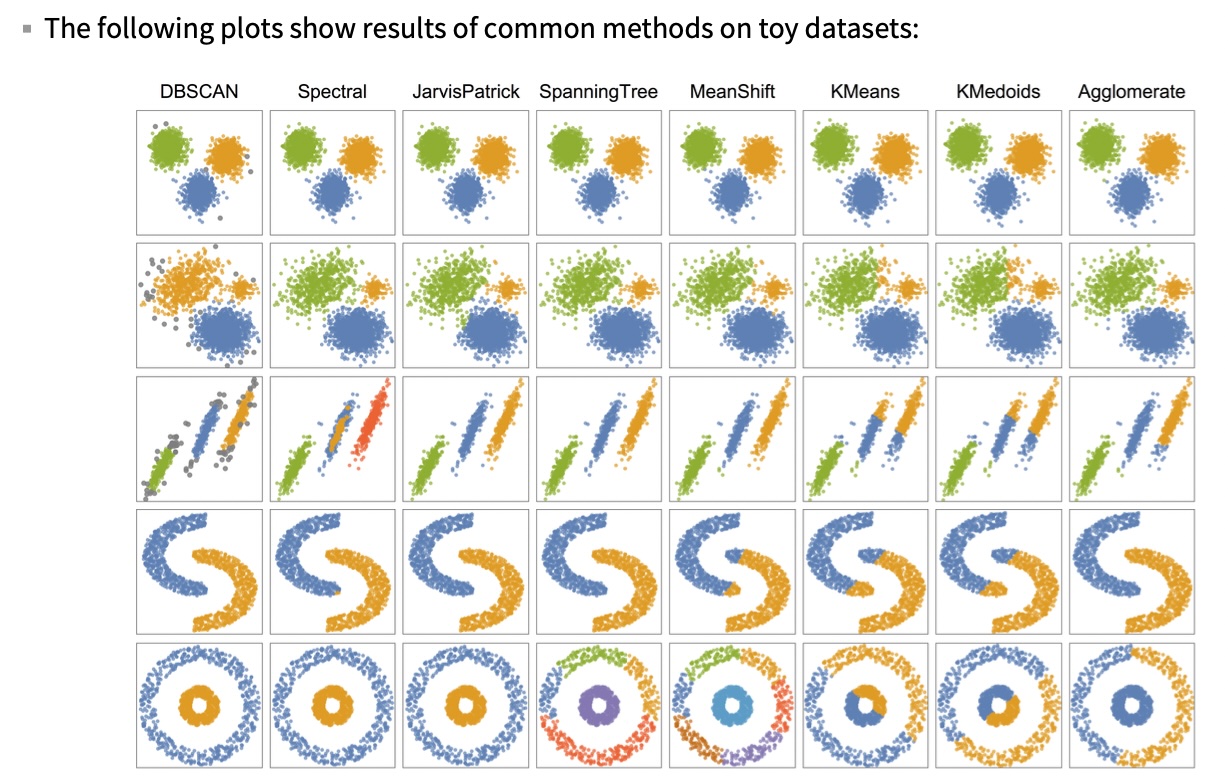
Clustering Methods Comparison
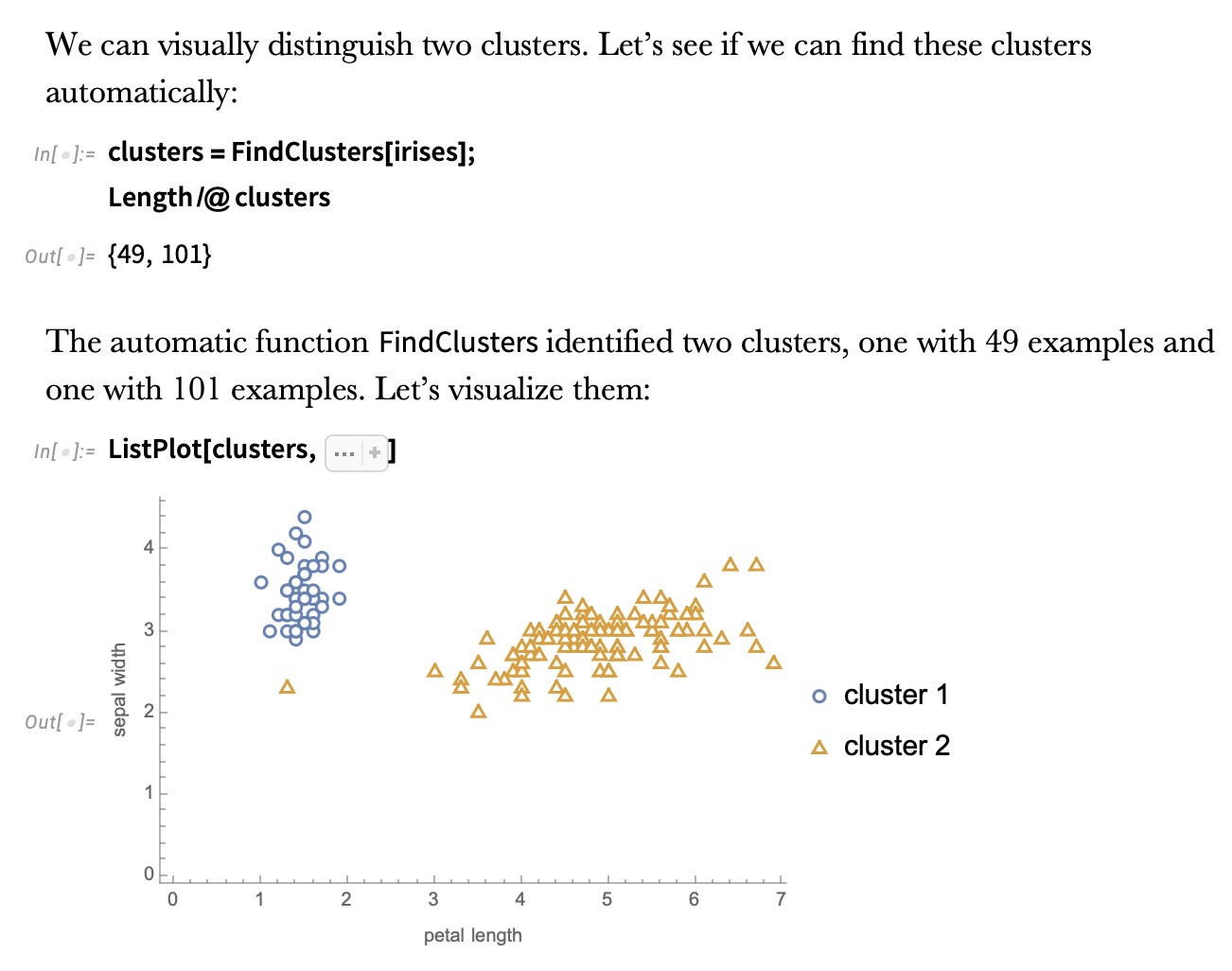
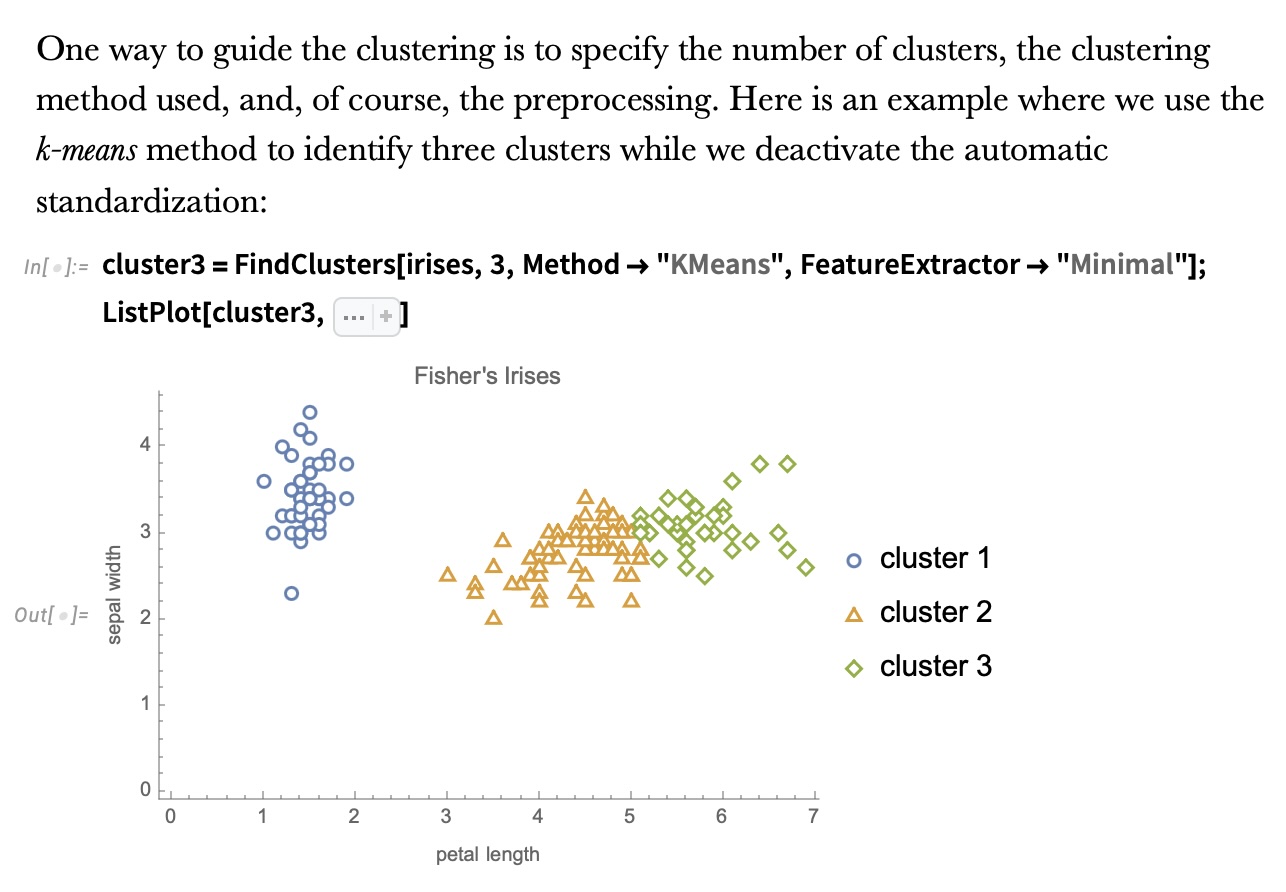
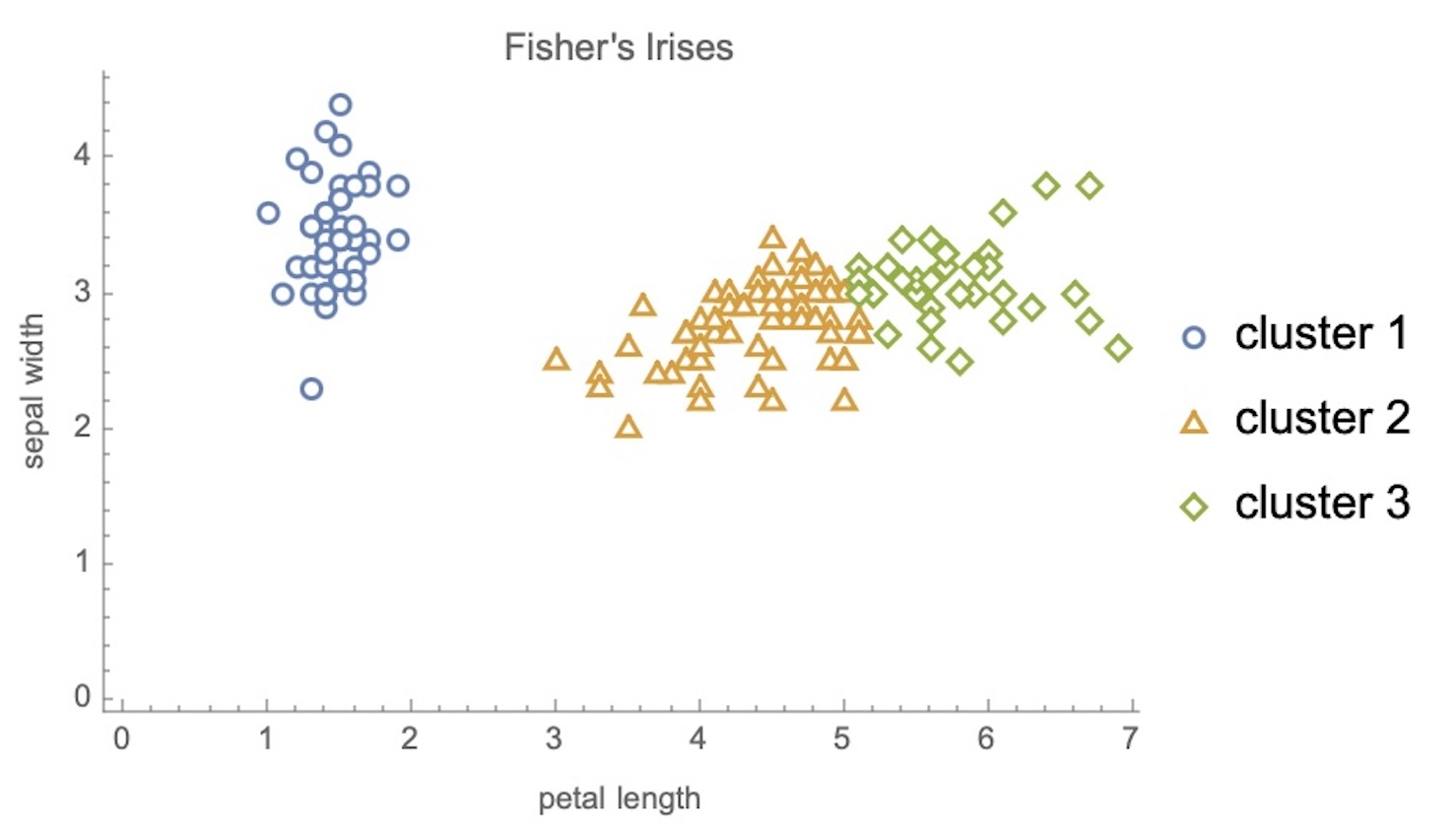
K-means Clustering
Clustering Method Zoo
Visual Comparison of Methods
Ground Truth vs K-means
Dimensionality Reduction
The Challenge: High-Dimensional Data
- Real-world data often has hundreds or thousands of dimensions
- Images: thousands of pixels
- Text: thousands of words
- Urban sensors: hundreds of measurements
- Customer data: dozens to hundreds of features
- Problem: We can only visualize 2D or 3D!
The Manifold Hypothesis
- Key insight: High-dimensional data often lies on lower-dimensional structures
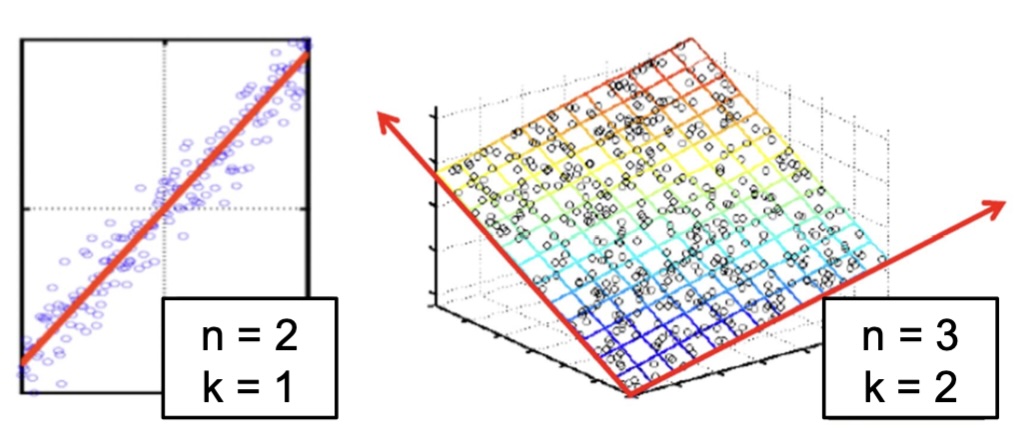
Non-linear Manifolds
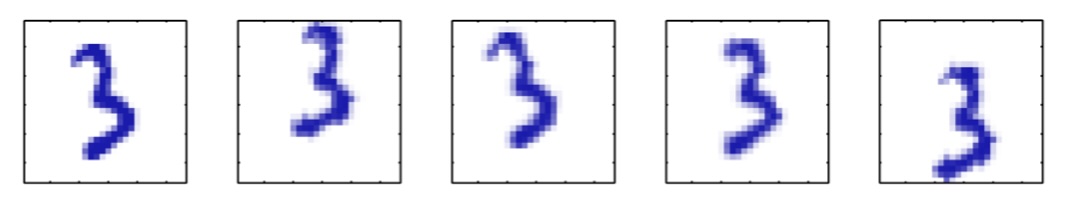
- Images of ‘3’ transformed by rotation, scaling, translation
- What’s the intrinsic dimensionality? (~5-7 dimensions)
- The underlying manifold is non-linear
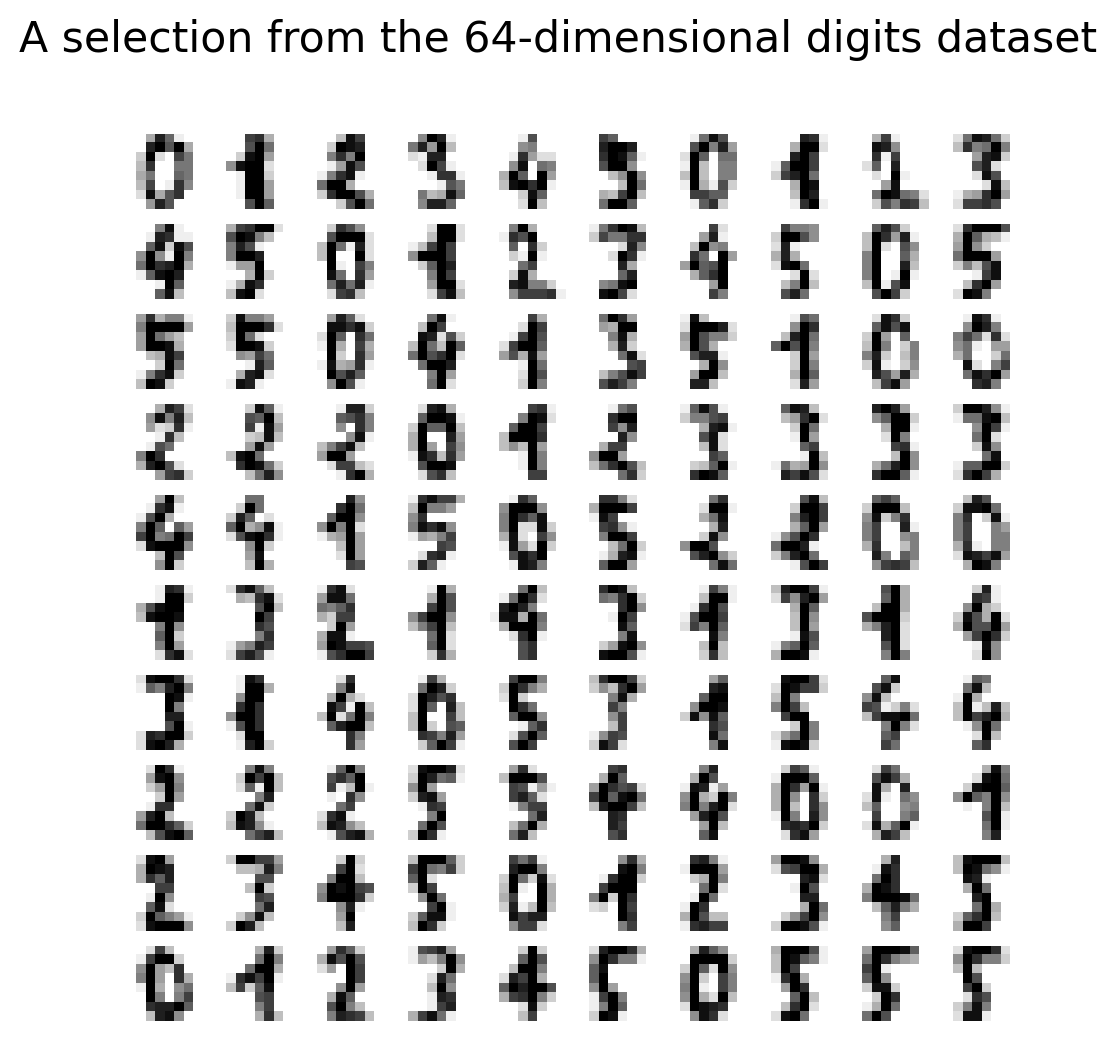
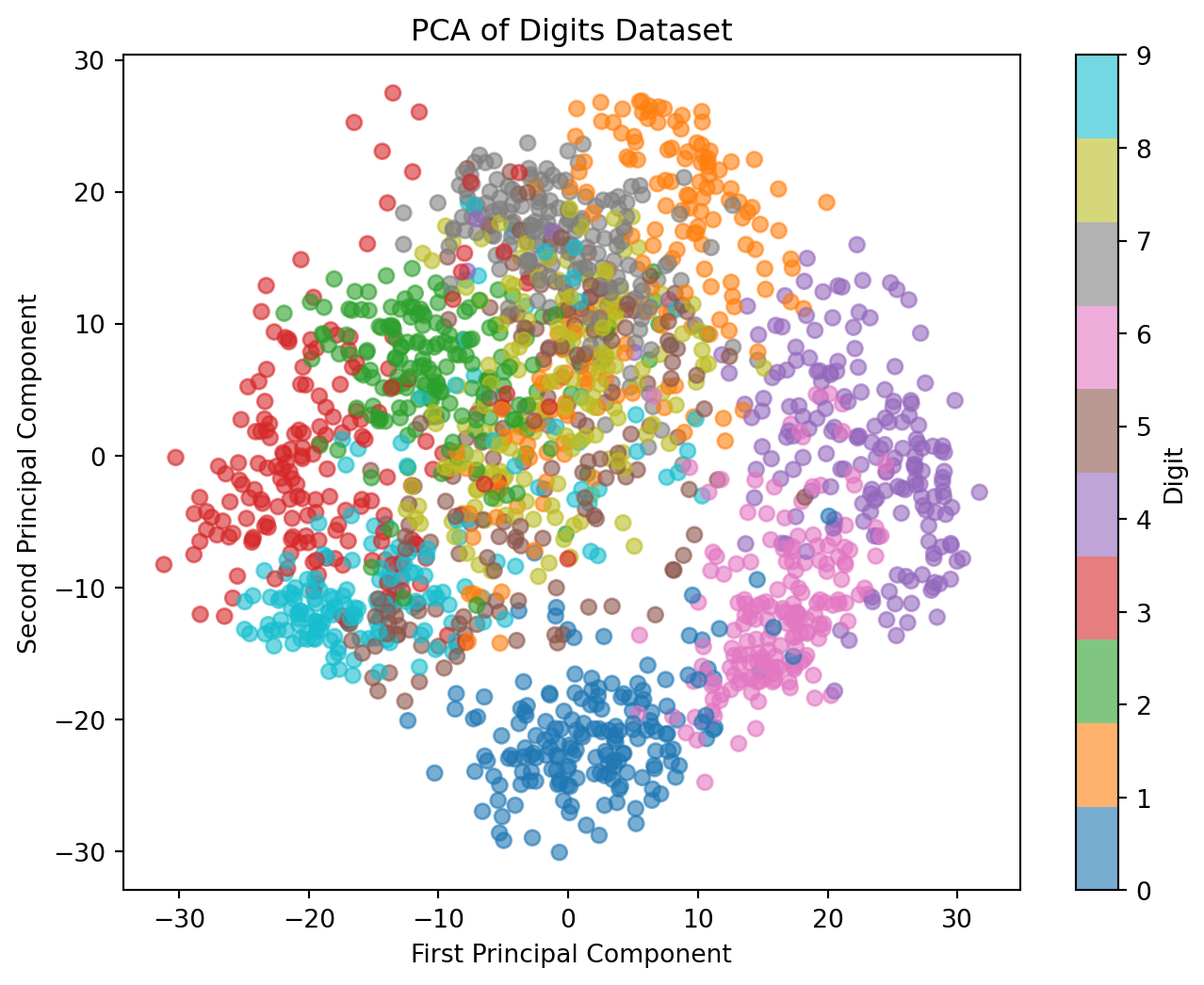
Working Example: Digits Dataset
- 8×8 pixel images = 64 dimensions
- Can’t visualize 64D directly
- Goal: Create meaningful 2D visualization
Digits Dataset Sample
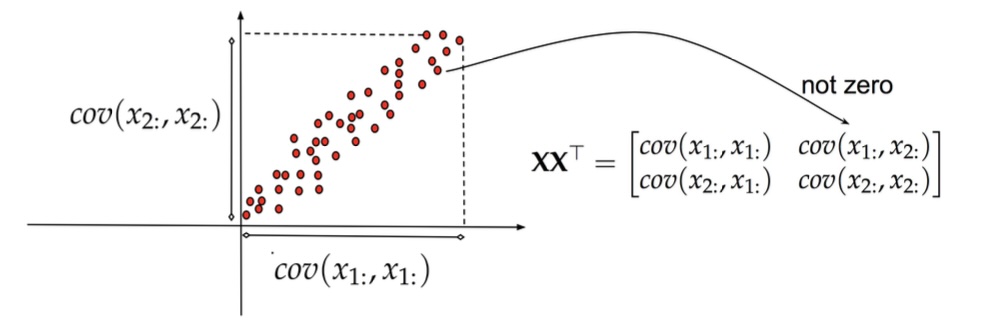
Principal Component Analysis (PCA)
PCA: The Foundation
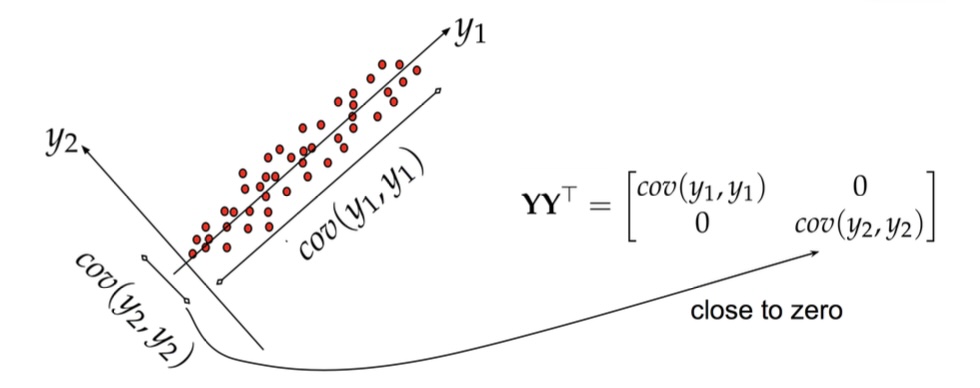
- PCA finds directions of maximum variance in data
- These directions are the eigenvectors of the covariance matrix
- Projects data onto these principal directions
- Linear method - assumes linear relationships
PCA: Geometric Intuition
PCA: Another View
PCA Applied to Digits
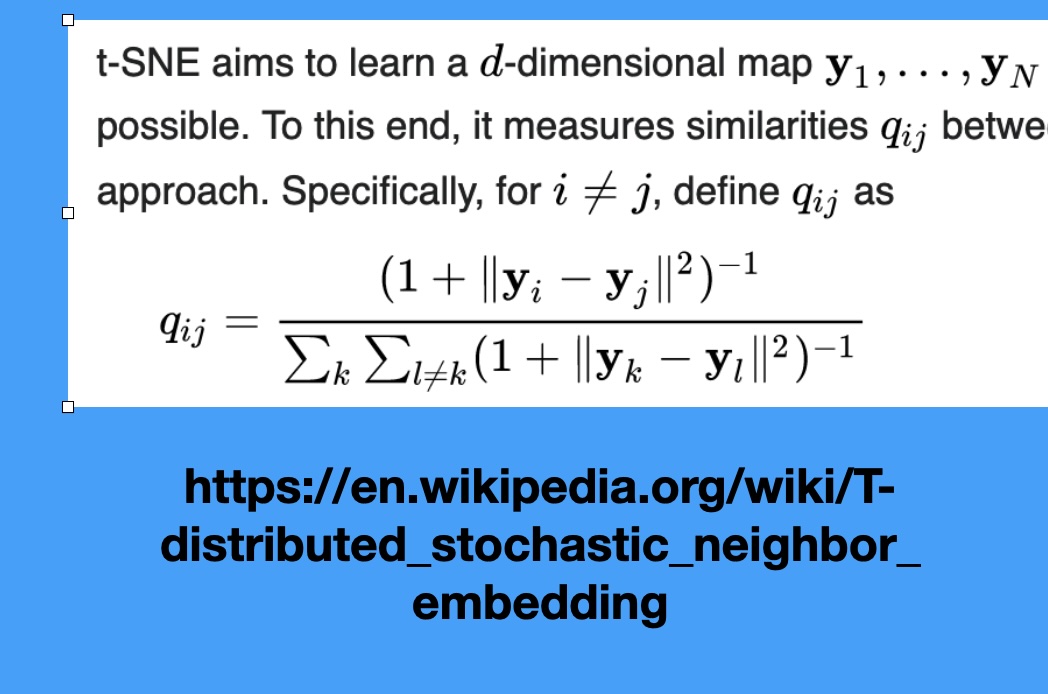
Non-linear Methods: t-SNE
t-SNE: A Revolution in Visualization
- t-SNE (t-Distributed Stochastic Neighbor Embedding)
- Preserves local neighborhood structure
- Non-linear, adapts to different regions of data
- Creates compelling cluster visualizations
- Warning: Can be easily misread!
How t-SNE Works

Critical Resource: “How to Use t-SNE Effectively”
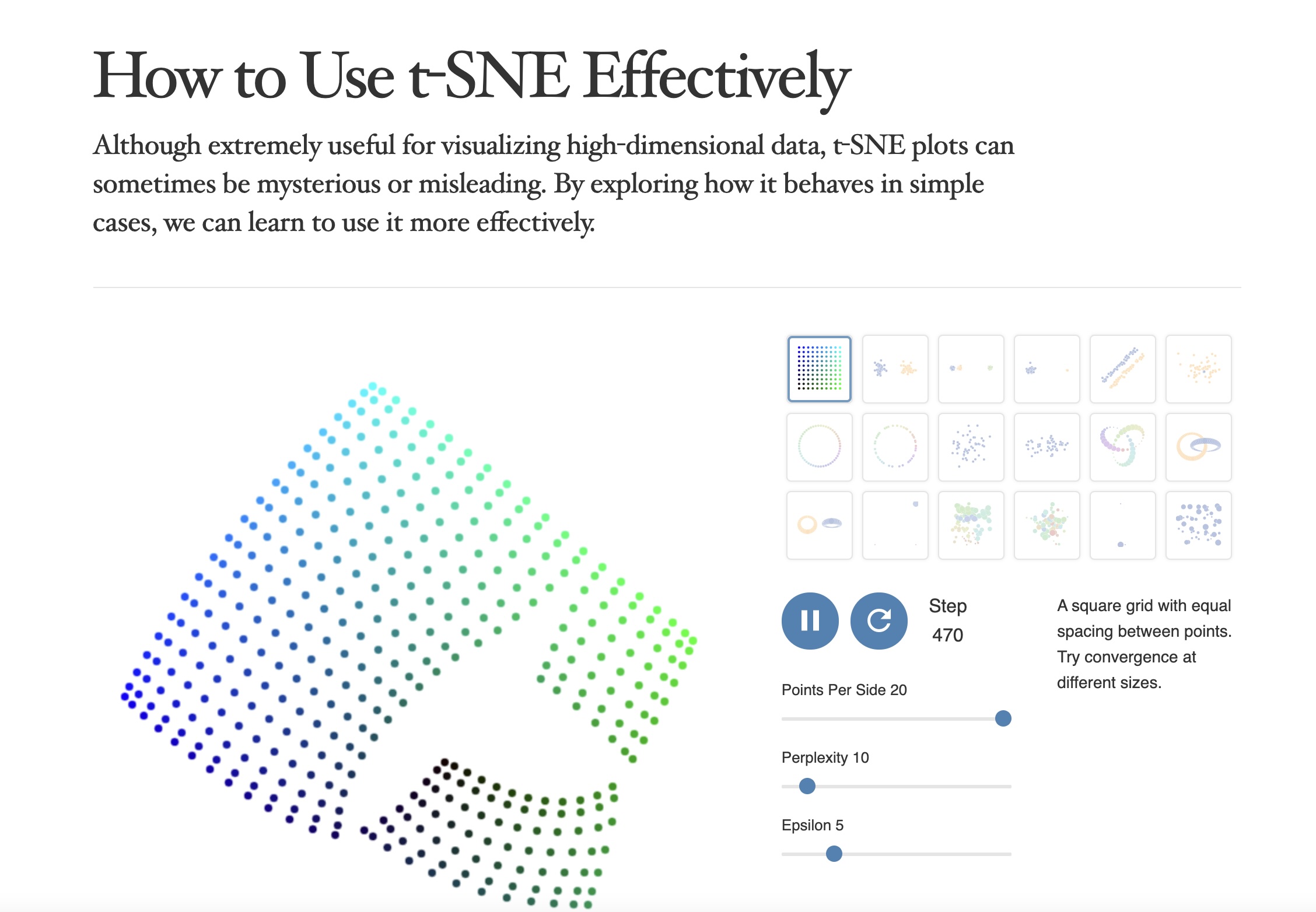
https://distill.pub/2016/misread-tsne/
Key Warning: Parameters Matter!
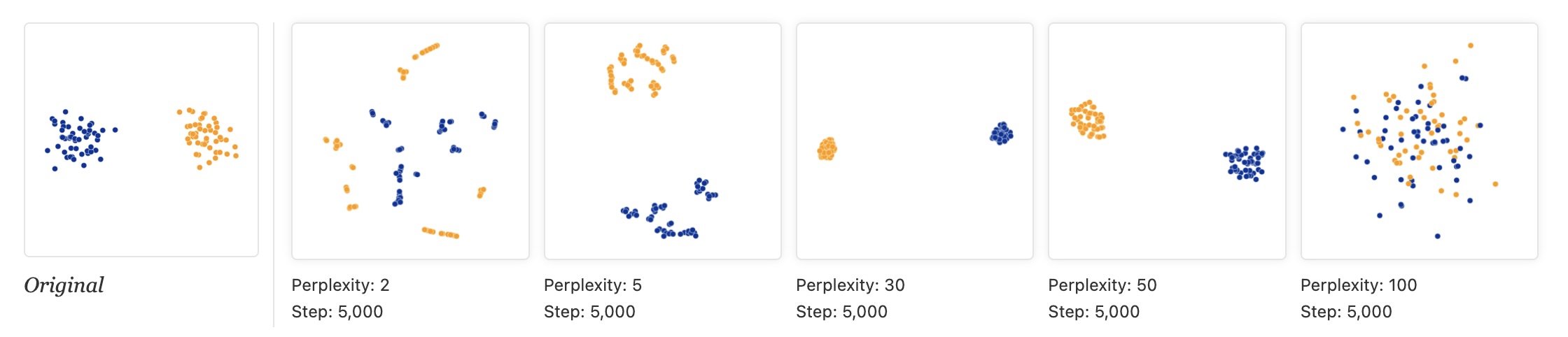
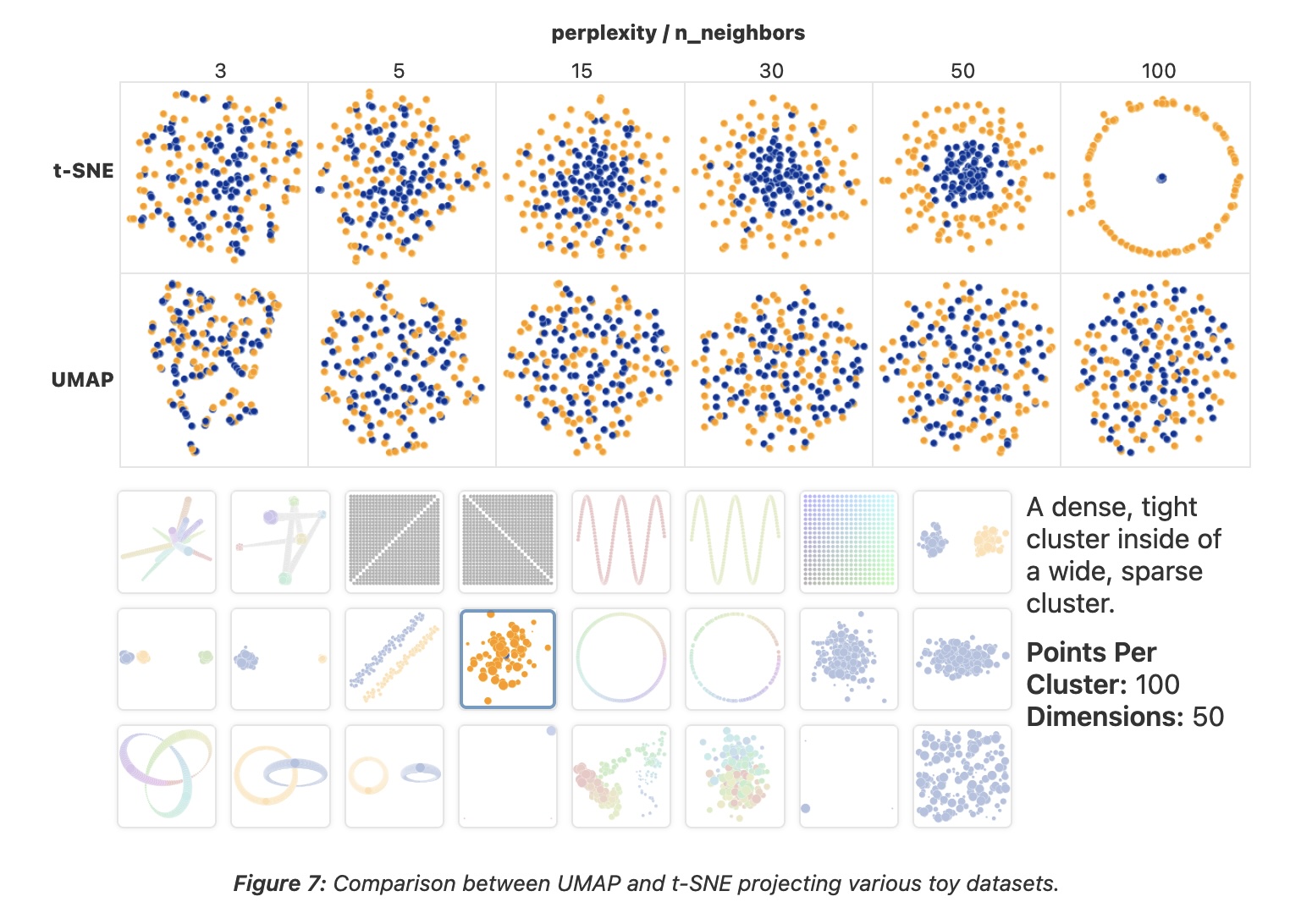
- t-SNE has a critical parameter: perplexity
- Roughly the number of close neighbors each point has
- Typical values: 5-50
- Different perplexities = different structures!
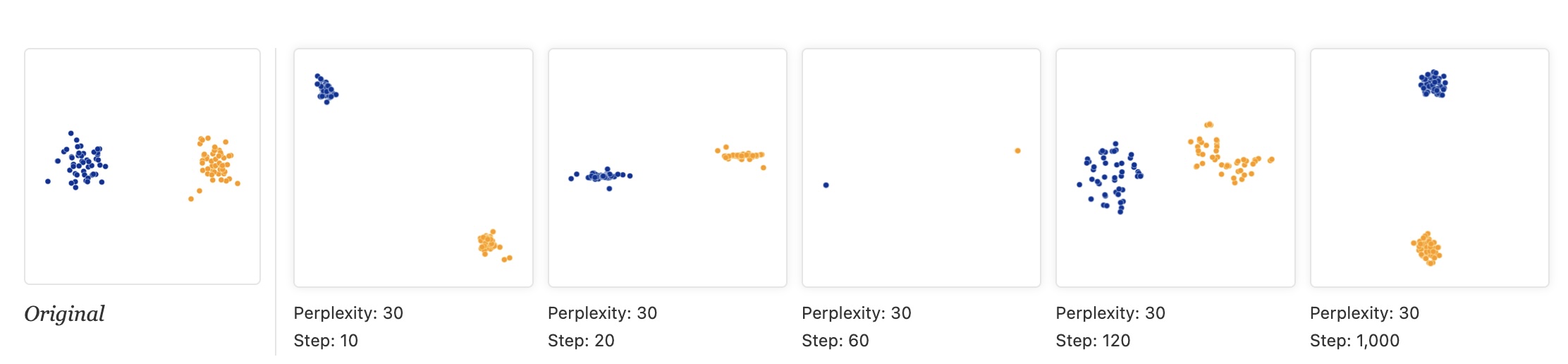
Convergence: Run Enough Iterations!
Critical: Cluster Sizes Mean Nothing!
- t-SNE equalizes cluster densities
- Large visual clusters ≠ large actual clusters
- Size refers to spatial extent, not number of points
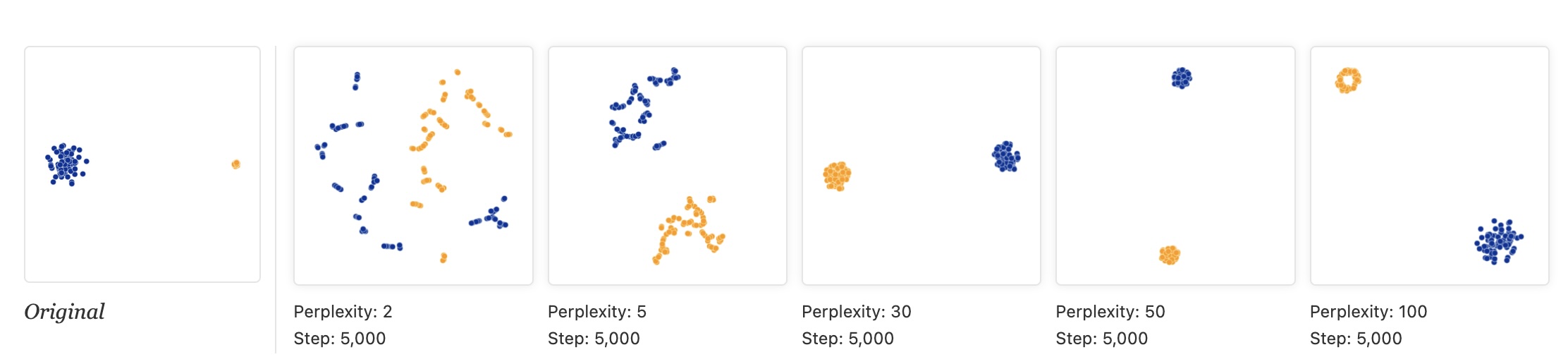
Critical: Distances Between Clusters Mean Nothing!

Warning: Random Noise Can Look Structured
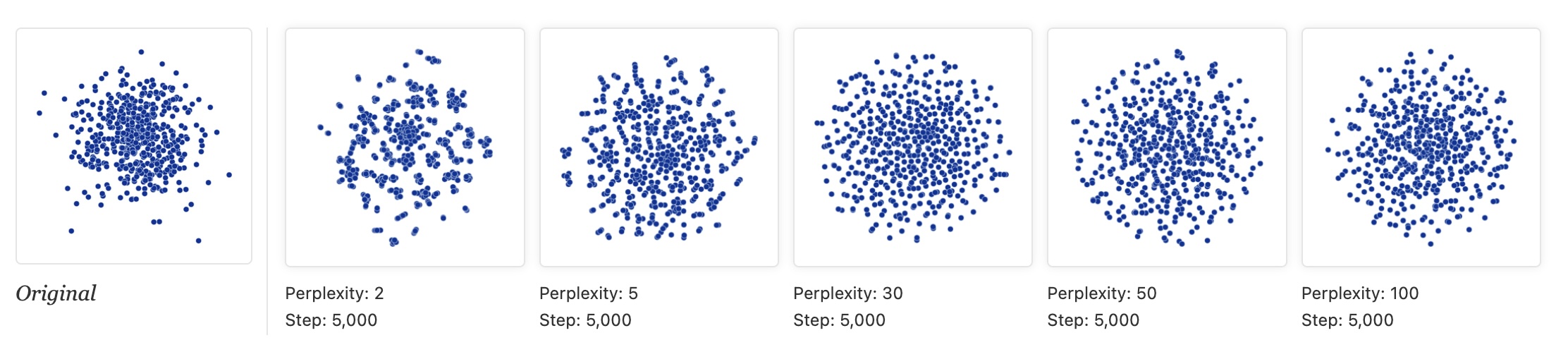
- Left: PCA of random data (correctly shows no structure)
- Right: t-SNE of same data (shows apparent clusters!)
- Lesson: Don’t assume clusters in t-SNE are real!
t-SNE Applied to Digits
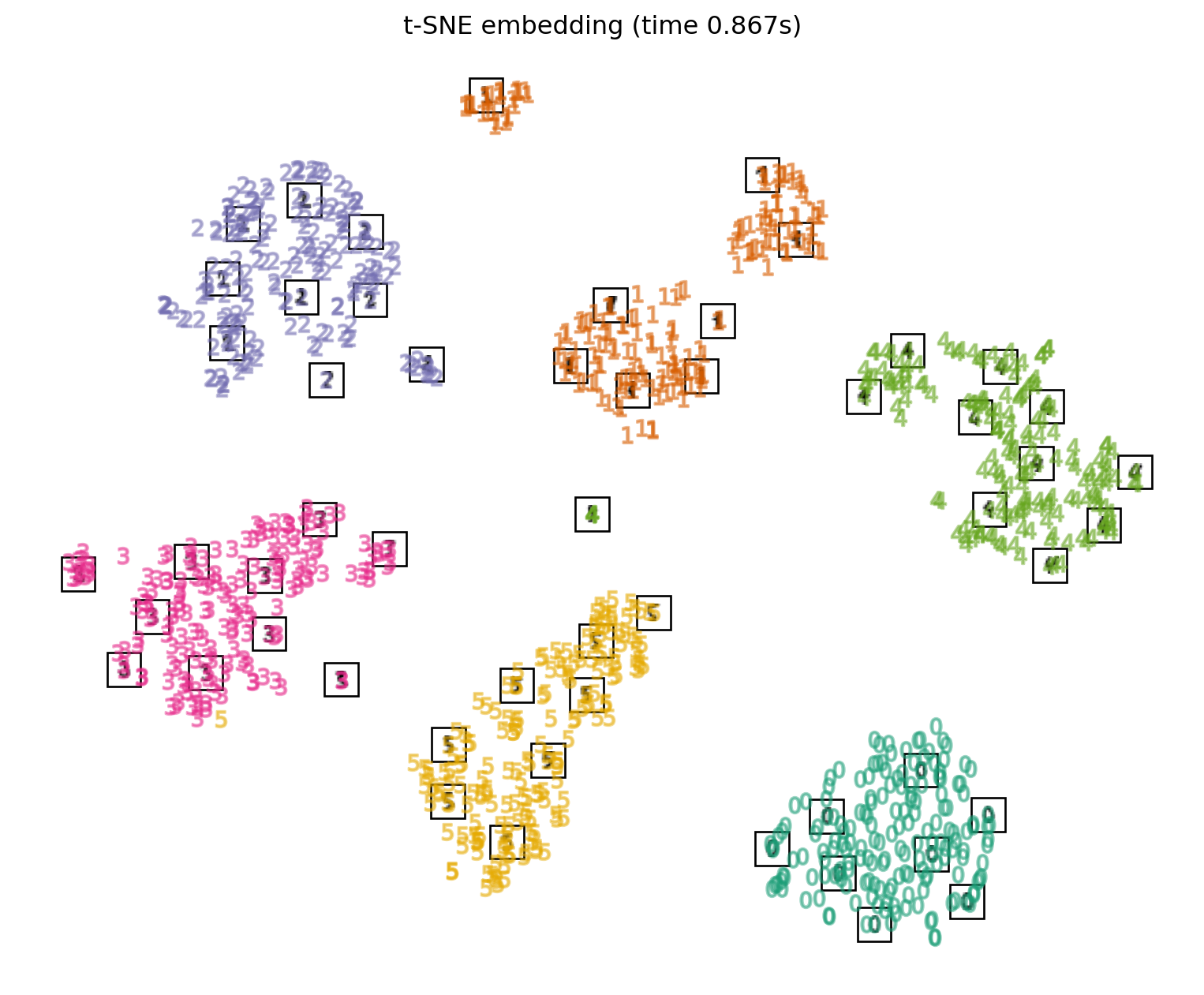
Non-linear Methods: UMAP
UMAP: The Modern Alternative
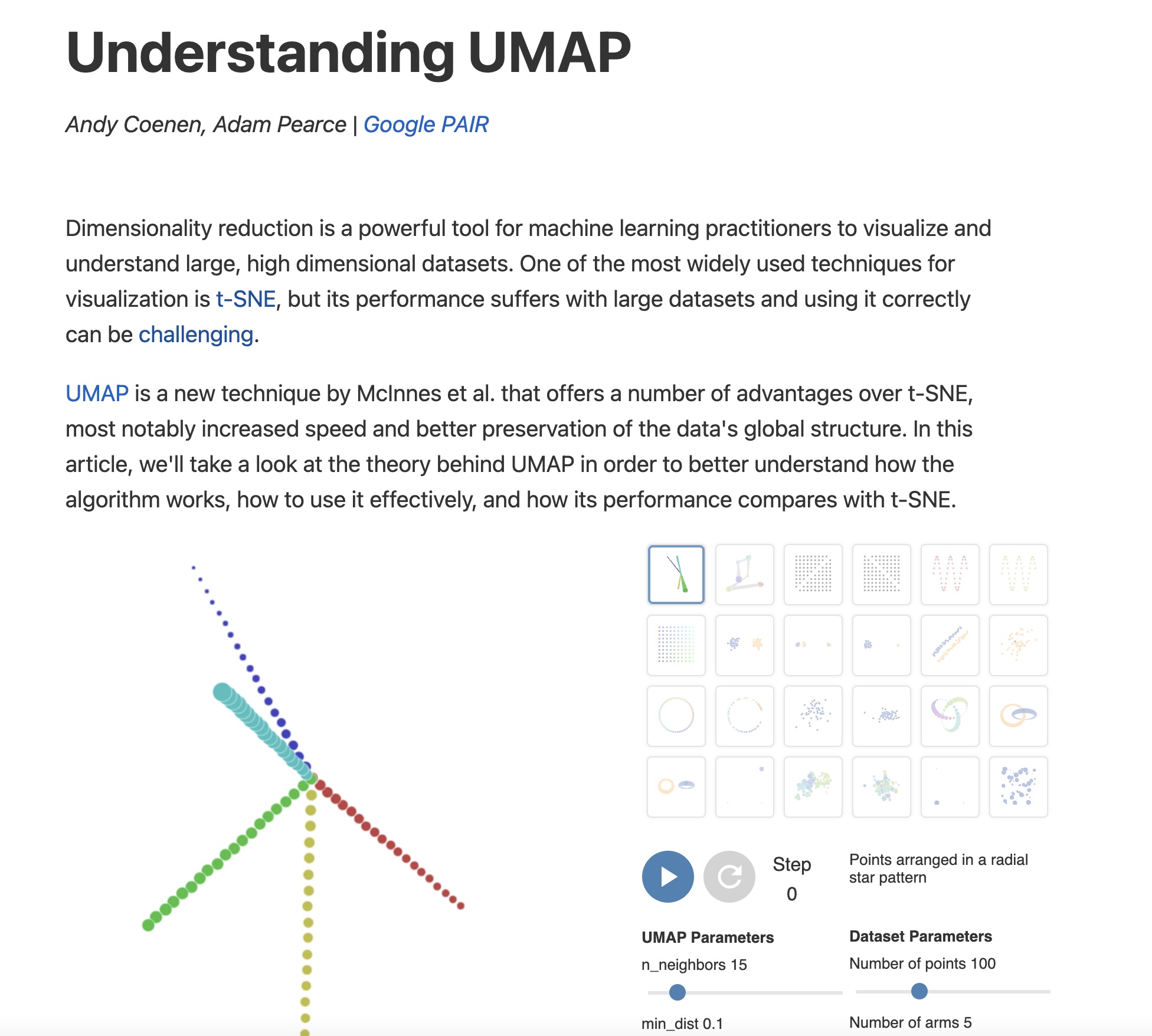
https://pair-code.github.io/understanding-umap/
UMAP vs t-SNE: Speed and Structure
- Speed: UMAP is 10-15x faster than t-SNE
- MNIST (70K points, 784D): UMAP = 3 min, t-SNE = 45 min!
- Global structure: UMAP better preserves relationships between clusters
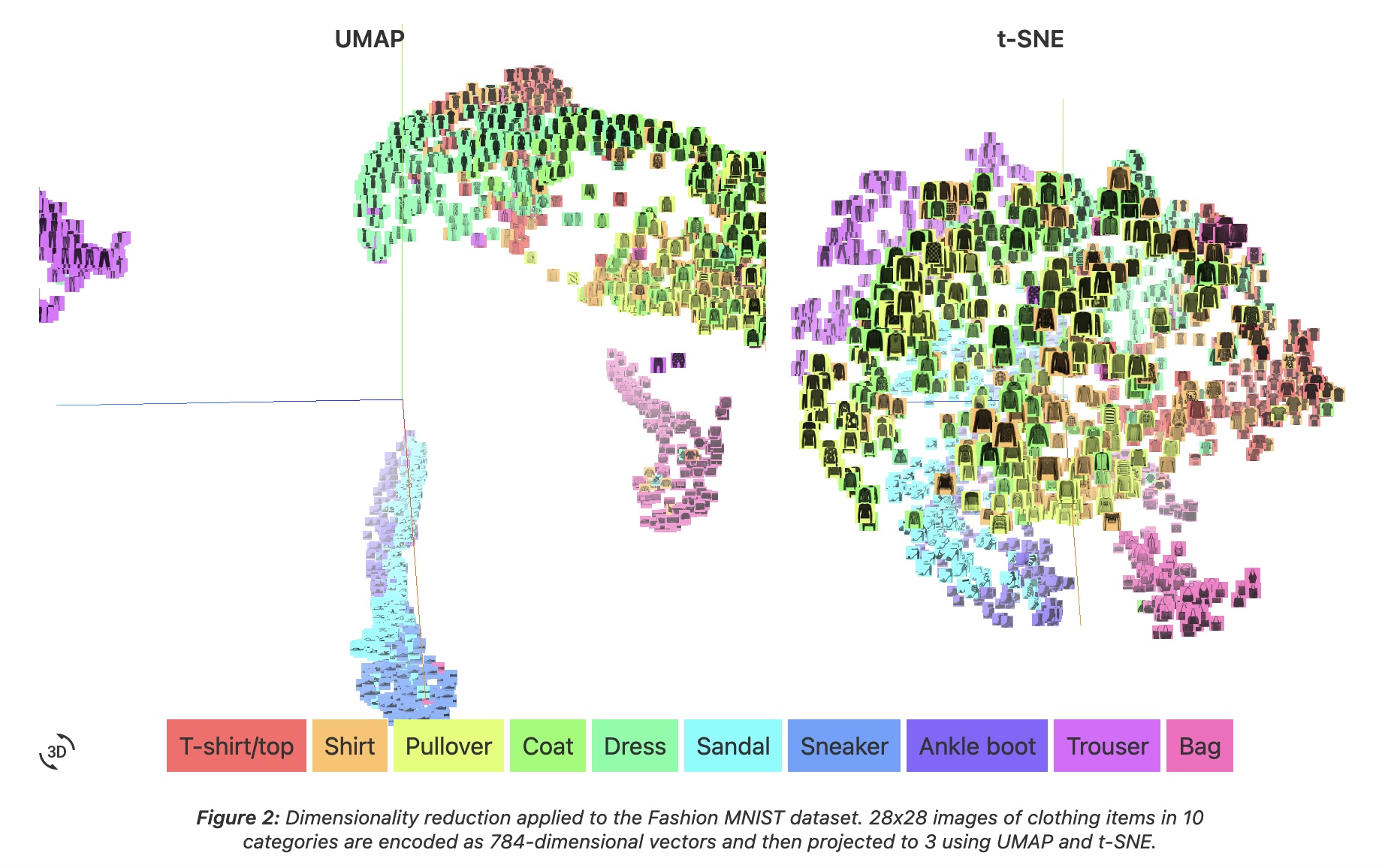
How UMAP Works
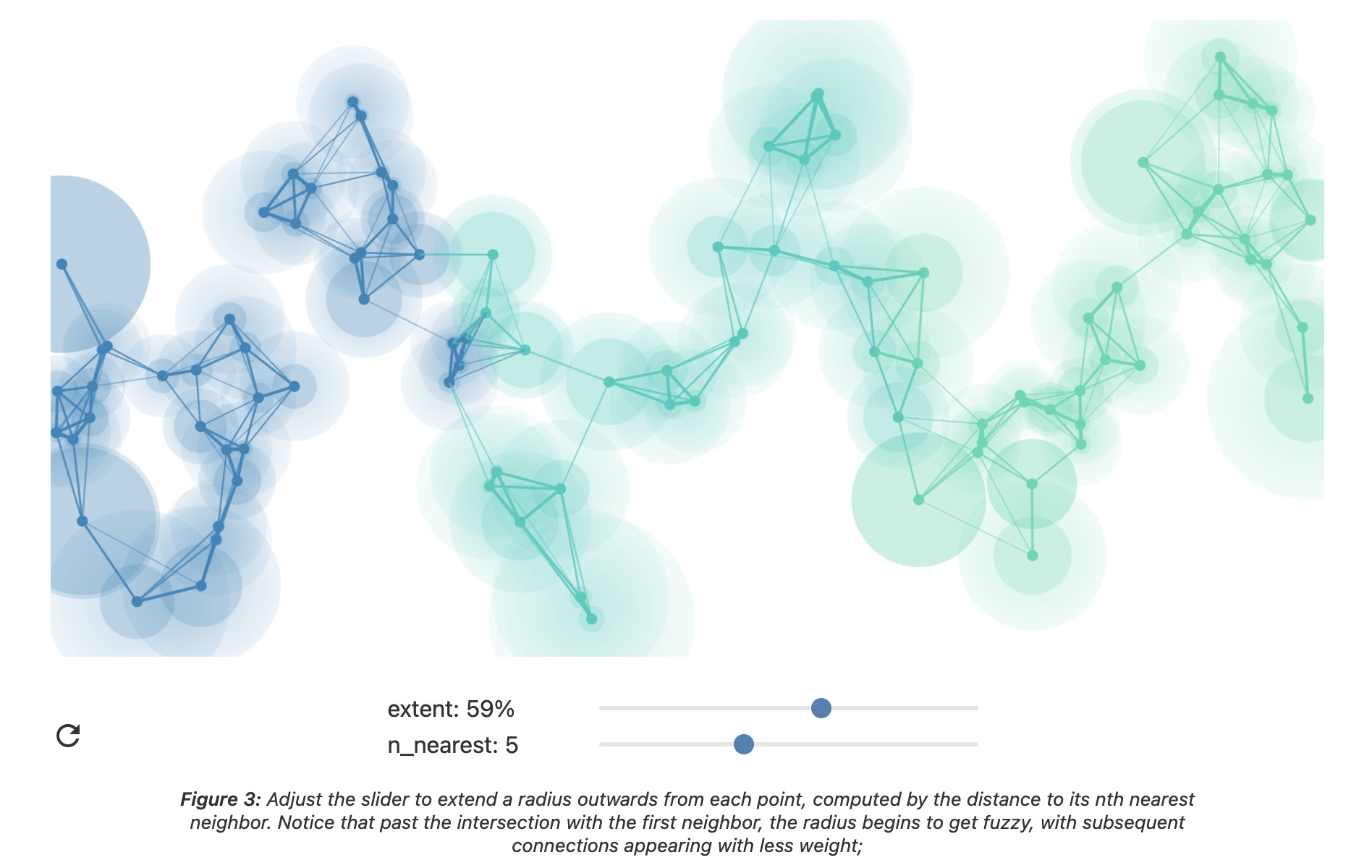
UMAP Parameters
- n_neighbors - balances local vs global structure
- Low (2-5): very local structure
- High (50-200): more global structure
- Similar to t-SNE’s perplexity
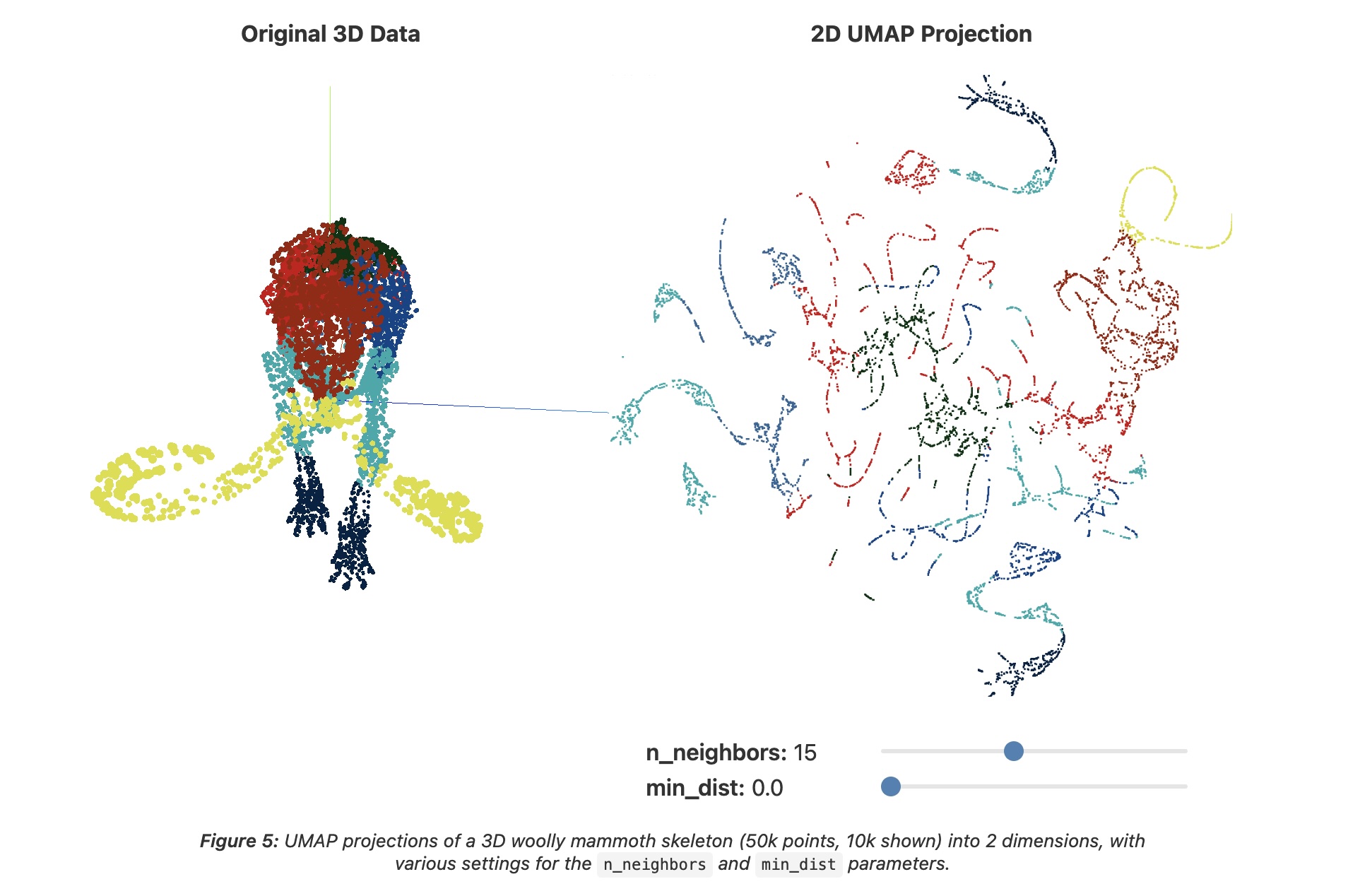
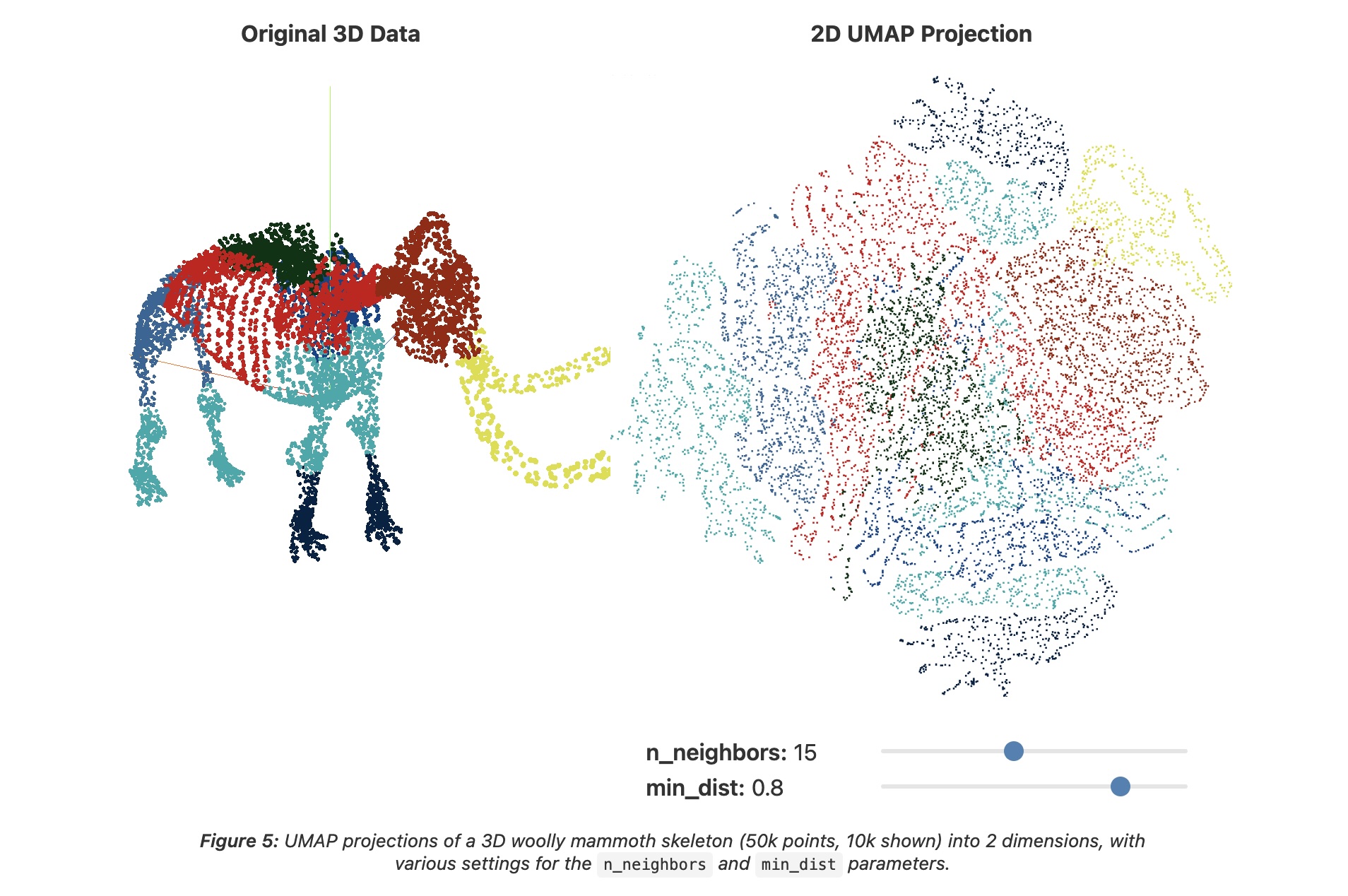
- min_dist - minimum spacing between points
- Low (0.0): tight clusters
- High (0.5-0.8): spread out points
UMAP: min_dist = 0.0
UMAP: min_dist = 0.8
UMAP: Critical Warnings Still Apply!
- UMAP is non-linear like t-SNE
- Distances are not directly interpretable
- Global positioning is better than t-SNE but still not perfect
- Always try multiple parameter values
- Validate findings with domain knowledge
Parameter Comparison: t-SNE vs UMAP
Visualization Best Practices
Dimensionality Reduction: Visualization Guidelines
- Always show multiple views
- PCA (linear baseline)
- t-SNE or UMAP (non-linear)
- Try multiple parameters
- Don’t over-interpret
- Cluster sizes may be misleading
- Inter-cluster distances may be unreliable
- Some structure may be artifacts
- Validate findings
- Check against domain knowledge
- Use complementary analyses
- Be transparent about limitations
Interactive Dimensionality Reduction
Summary
- Clustering helps discover natural groups in data
- Multiple methods reveal different structures
- Visualization is essential for interpretation
- Dimensionality Reduction enables visualization of complex data
- PCA: fast, interpretable, linear
- t-SNE: excellent clusters, requires care
- UMAP: faster, better global structure
- Critical: Always validate, try multiple methods, understand limitations
Resources and Further Reading
Clustering: * Wolfram Clustering Tutorial * Cluster Analysis (Wikipedia)
Dimensionality Reduction: * How to Use t-SNE Effectively (Essential!) * Understanding UMAP * Dimensionality Reduction (Wikipedia)
